4WEB
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMAMIDE, Mouse Fab Heavy Chain, ... | | Authors: | Khan, A.G, Whidby, J, Miller, M.T, Scarborough, H, Zatorski, A.V, Cygan, A, Price, A.A, Yost, S.A, Bohannon, C.D, Jacob, J, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2.
Nature, 509, 2014
|
|
6XI7
 
 | | Crystal Structure of wild-type KRAS (GMPPNP-bound) in complex with RAS-binding domain (RBD) and cysteine-rich domain (CRD) of RAF1/CRAF (crystal form I) | | Descriptor: | CHLORIDE ION, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Chan, A.H, Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-06-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
7N0Z
 
 | |
4WCM
 
 | |
4WCL
 
 | |
4WCN
 
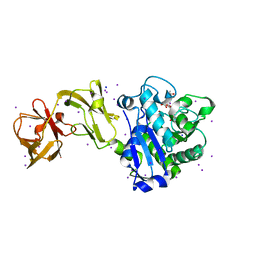 | | Crystal Structure of Tripeptide bound Cell Shape Determinant Csd4 protein from Helicobacter pylori | | Descriptor: | Conserved hypothetical secreted protein, IODIDE ION, N-acetyl-L-alanyl-N-[(1S,5R)-5-amino-1,5-dicarboxypentyl]-D-glutamine, ... | | Authors: | Chan, A.C, Murphy, M.E. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Helical Shape of Helicobacter pylori Requires an Atypical Glutamine as a Zinc Ligand in the Carboxypeptidase Csd4.
J.Biol.Chem., 290, 2015
|
|
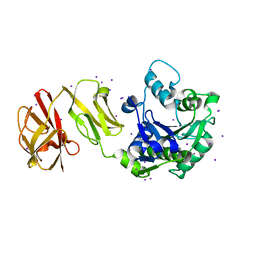
4WCK
 
 | |
1ELN
 
 | |
5VQV
 
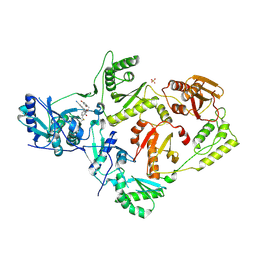 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQQ
 
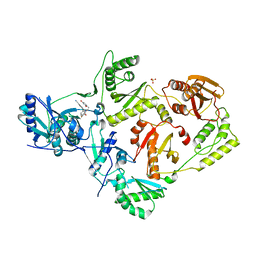 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide (JLJ683), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-2-fluoro-N-methylacetamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VQY
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) Variant in Complex with N-(6-cyano-3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-4-methylnaphthalen-1-yl)-N-methylacrylamide (JLJ684), a Non-nucleoside Inhibitor | | Descriptor: | N-(6-cyano-3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-4-methylnaphthalen-1-yl)-N-methylpropanamide, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Anderson, K.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Covalent inhibitors for eradication of drug-resistant HIV-1 reverse transcriptase: From design to protein crystallography.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7Z2J
 
 | | White Bream virus N7-Methyltransferase | | Descriptor: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
3VNE
 
 | | Structure of the ebolavirus protein VP24 from Sudan | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
4PPQ
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-CR | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
2G5G
 
 | |
4PR0
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-N3 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PSA
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-N1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PO9
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-BR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4POA
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-BN | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PPF
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-N | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PPG
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-BR | | Descriptor: | CITRATE ANION, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PQY
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-N4 | | Descriptor: | GLYCEROL, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | Descriptor: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4PO8
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-CR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PQF
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIB-CR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-03 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
