5M99
 
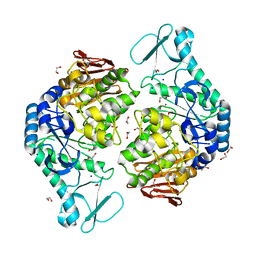 | | Functional Characterization and Crystal Structure of Thermostable Amylase from Thermotoga petrophila, reveals High Thermostability and an Archaic form of Dimerization | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-amylase, ... | | Authors: | Hameed, U, Price, I, Mirza, O.A. | | Deposit date: | 2016-11-01 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional characterization and crystal structure of thermostable amylase from Thermotoga petrophila, reveals high thermostability and an unusual form of dimerization.
Biochim. Biophys. Acta, 1865, 2017
|
|
8J2S
 
 | |
5Z95
 
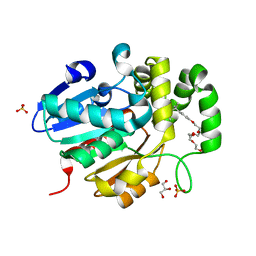 | | Structural basis for specific inhibition of highly sensitive ShHTL7 receptor | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Hameed, U.S, Arold, S.T. | | Deposit date: | 2018-02-02 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor.
EMBO Rep., 19, 2018
|
|
5Z82
 
 | |
5Z89
 
 | | Structural basis for specific inhibition of highly sensitive ShHTL7 receptor | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Hameed, U.S, Arold, S.T. | | Deposit date: | 2018-01-31 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor.
EMBO Rep., 19, 2018
|
|
5Z8P
 
 | |
6LF3
 
 | |
7W5C
 
 | | Crystal structure of Mitogen Activated Protein Kinase 4 (MPK4) from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 4, Mitogen-activated protein kinase kinase 1, ... | | Authors: | Arold, S.T, Hameed, U.F.S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Essential role of the CD docking motif of MPK4 in plant immunity, growth, and development.
New Phytol., 239, 2023
|
|
7C8L
 
 | | Hybrid designing of potent inhibitors of Striga strigolactone receptor ShHTL7 | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Shahul Hameed, U.F, Arold, S.T. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of Striga hermonthica-specific seed germination inhibitors.
Plant Physiol., 188, 2022
|
|
6LES
 
 | |
6IPZ
 
 | | Fyn SH3 domain R96W mutant, crystallized with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Tyrosine-protein kinase Fyn | | Authors: | Arold, S.T, Aljedani, S.S, Shahul Hameed, U.F. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
6IPY
 
 | |
8J31
 
 | |
8J2V
 
 | |
8J2U
 
 | |
5ZTZ
 
 | |
5ZU0
 
 | |
5ZZW
 
 | |
8J2T
 
 | |
8J2Z
 
 | |
7D7S
 
 | |
