1XWE
 
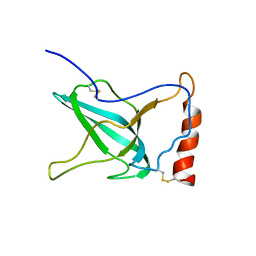 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
3EC1
 
 | |
6EA6
 
 | |
1F8F
 
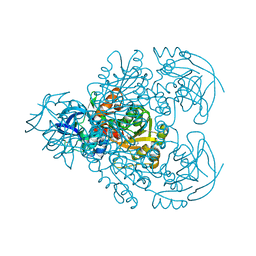 | | CRYSTAL STRUCTURE OF BENZYL ALCOHOL DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | Descriptor: | BENZYL ALCOHOL DEHYDROGENASE, ETHANOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Beauchamp, J.C, Gillooly, D, Warwicker, J, Fewson, C.A, Lapthorn, A.J. | | Deposit date: | 2000-06-30 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Benzyl Alcohol Dehydrogenase from Acinetobacter calcoaceticus
To be Published
|
|
8UA4
 
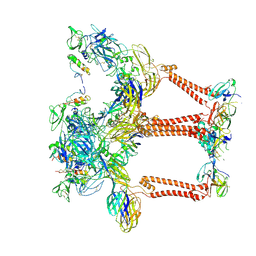 | | Structure of eastern equine encephalitis virus VLP in complex with VLDLR LA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid protein, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
6XB3
 
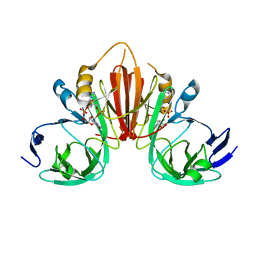 | |
8UA9
 
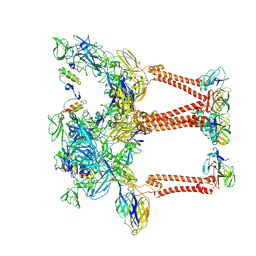 | | Structure of eastern equine encephalitis virus VLP unliganded quasi-threefold spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, Envelope glycoprotein E1, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for VLDLR recognition by eastern equine encephalitis virus.
Nat Commun, 15, 2024
|
|
8UA8
 
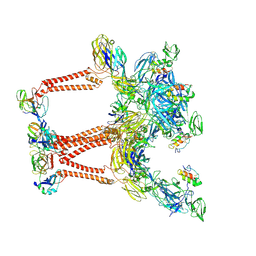 | | Structure of Semliki Forest virus VLP in complex with VLDLR LA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Assembly protein E3, ... | | Authors: | Abraham, J, Yang, P, Li, W, Fan, X, Pan, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of SFV VLP-VLDLRAD2 complex at the 3-fold axes
To Be Published
|
|
1UZ9
 
 | | Crystallographic and solution studies of N-lithocholyl insulin: a new generation of prolonged-acting insulins. | | Descriptor: | (2S)-2-AMINO-6-({(4R)-4-[(10R,13S)-10,13-DIMETHYL-3-OXOHEXADECAHYDRO-1H-CYCLOPENTA[A]PHENANTHREN-17-YL]PENTANOYL}AMINO)HEXANOIC ACID, CHLORIDE ION, INSULIN, ... | | Authors: | Whittingham, J.L, Jonassen, I, Havelund, S, Roberts, S.M, Dodson, E.J, Verma, C.S, Wilkinson, A.J, Dodson, G.G. | | Deposit date: | 2004-03-08 | | Release date: | 2005-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and Solution Studies of N-Lithocholyl Insulin: A New Generation of Prolonged-Acting Human Insulins
Biochemistry, 43, 2004
|
|
1TTA
 
 | |
1XDA
 
 | | STRUCTURE OF INSULIN | | Descriptor: | CHLORIDE ION, FATTY ACID ACYLATED INSULIN, MYRISTIC ACID, ... | | Authors: | Whittingham, J.L, Havelund, S, Jonassen, I. | | Deposit date: | 1996-12-18 | | Release date: | 1997-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a prolonged-acting insulin with albumin-binding properties.
Biochemistry, 36, 1997
|
|
1TTC
 
 | |
6EA9
 
 | |
5C5U
 
 | | The crystal structure of viral collagen prolyl hydroxylase vCPH from Paramecium Bursaria Chlorella virus-1 - Truncated Construct | | Descriptor: | ACETATE ION, MANGANESE (II) ION, Prolyl 4-hydroxylase, ... | | Authors: | Longbotham, J.E, Levy, C.W, Johannisen, L.O, Tarhonskaya, H, Jiang, S, Loenarz, C, Flashman, E, Hay, S, Schofiled, C.J, Scrutton, N.S. | | Deposit date: | 2015-06-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of a Viral Collagen Prolyl Hydroxylase.
Biochemistry, 54, 2015
|
|
1HEW
 
 | |
1H67
 
 | | NMR Structure of the CH Domain of Calponin | | Descriptor: | CALPONIN ALPHA | | Authors: | Bramham, J, Smith, B.O, Uhrin, D, Barlow, P.N, Winder, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2002-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Calponin Ch Domain and Fitting to the 3D-Helical Reconstruction of F-Actin:Calponin.
Structure, 10, 2002
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
1MPJ
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | CHLORIDE ION, PHENOL, PHENOL INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
2CEU
 
 | | Despentapeptide insulin in acetic acid (pH 2) | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Zhang, Y, Zakova, L, Dodson, E.J, Turkenburg, J.P, Brange, J, Dodson, G.G. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | I222 Crystal Form of Despentapeptide (B26-B30) Insulin Provides New Insights Into the Properties of Monomeric Insulin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3MTH
 
 | | X-RAY CRYSTALLOGRAPHIC STUDIES ON HEXAMERIC INSULINS IN THE PRESENCE OF HELIX-STABILIZING AGENTS, THIOCYANATE, METHYLPARABEN AND PHENOL | | Descriptor: | 4-HYDROXY-BENZOIC ACID METHYL ESTER, CHLORIDE ION, METHYLPARABEN INSULIN, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Moody, P.C.E, Dodson, G.G. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystallographic studies on hexameric insulins in the presence of helix-stabilizing agents, thiocyanate, methylparaben, and phenol.
Biochemistry, 34, 1995
|
|
1GUJ
 
 | | Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation. | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Scott, D.J, Chance, K, Wilson, A, Finch, J, Brange, J, Dodson, G.G. | | Deposit date: | 2002-01-28 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Insulin at Ph2: Structural Analysis of the Conditions Promoting Insulin Fibre Formation
J.Mol.Biol., 318, 2002
|
|
4H1G
 
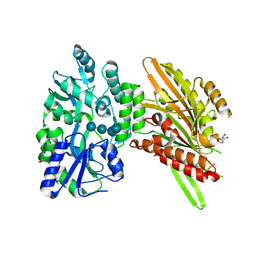 | | Structure of Candida albicans Kar3 motor domain fused to maltose-binding protein | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Allingham, J.A, Duan, D, Delorme, C, Joshi, M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein.
Biochem.Biophys.Res.Commun., 428, 2012
|
|
6T9M
 
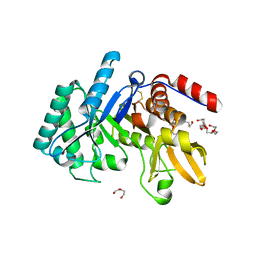 | | Crystal structure of the Chitinase Domain of the Spore Coat Protein CotE from Clostridium difficile | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptide in active site, ... | | Authors: | Whittingham, J.L, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of the GH18 domain of the bifunctional peroxiredoxin-chitinase CotE from Clostridium difficile.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TSB
 
 | |
