4UOB
 
 | | Crystal structure of Deinococcus radiodurans Endonuclease III-3 | | Descriptor: | ACETATE ION, COBALT (II) ION, ENDONUCLEASE III-3, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
4UNF
 
 | | Crystal structure of Deinococcus radiodurans Endonuclease III-1 | | Descriptor: | ACETATE ION, ENDONUCLEASE III-1, IRON/SULFUR CLUSTER, ... | | Authors: | Sarre, A, Okvist, M, Klar, T, Hall, D, Smalas, A.O, McSweeney, S, Timmins, J, Moe, E. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Two Unusual Endonuclease III Enzymes from Deinococcus Radiodurans.
J.Struct.Biol., 191, 2015
|
|
6WQU
 
 | | CSL (RBPJ) bound to Notch3 RAM and DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Neurogenic locus notch homolog protein 3, ... | | Authors: | Kovall, R.A, Gagliani, E, Hall, D. | | Deposit date: | 2020-04-29 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PIM-induced phosphorylation of Notch3 promotes breast cancer tumorigenicity in a CSL-independent fashion.
J.Biol.Chem., 296, 2021
|
|
3MSK
 
 | | Fragment Based Discovery and Optimisation of BACE-1 Inhibitors | | Descriptor: | 4-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-cyclohexyl-N-methylbutanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Smith, M.A, Madden, J.M, Barker, J, Godemann, R, Kraemer, J, Hallett, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1UHL
 
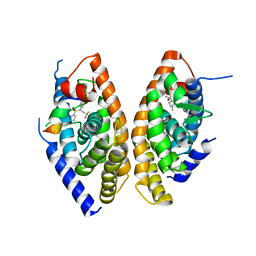 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
2EJN
 
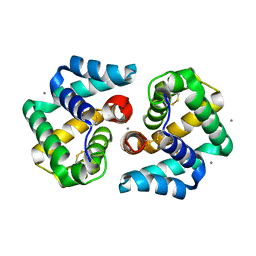 | | Structural characterization of the tetrameric form of the major cat allergen fel D 1 | | Descriptor: | CALCIUM ION, Major allergen I polypeptide chain 1, chain 2 | | Authors: | Kaiser, L, Velickovic, T.C, Badia-Martinez, D, Adedoyin, J, Thunberg, S, Hallen, D, Berndt, K, Gronlund, H, Gafvelin, G, van Hage, M, Achour, A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the tetrameric form of the major cat allergen Fel d 1
J.Mol.Biol., 370, 2007
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
7AEH
 
 | | SARS-CoV-2 main protease in a covalent complex with a pyridine derivative of ABT-957, compound 1 | | Descriptor: | (2~{R})-5-oxidanylidene-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]-1-(phenylmethyl)pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
1QW9
 
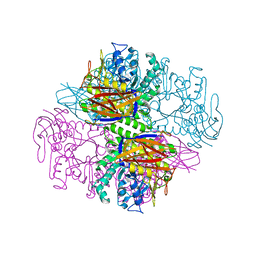 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with 4-nitrophenyl-Ara | | Descriptor: | 4-nitrophenyl alpha-L-arabinofuranoside, Alpha-L-arabinofuranosidase | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1QW8
 
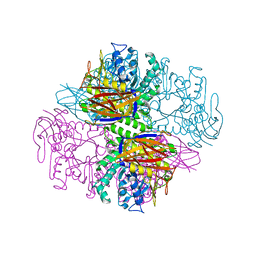 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1PZ2
 
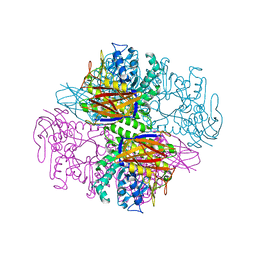 | | Crystal structure of a transient covalent reaction intermediate of a family 51 alpha-L-arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1PZ3
 
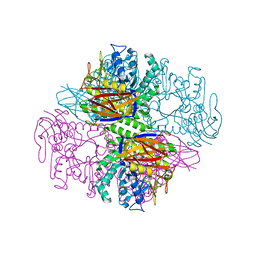 | | Crystal structure of a family 51 (GH51) alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T6 | | Descriptor: | Alpha-L-arabinofuranosidase, GLYCEROL | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
1K9E
 
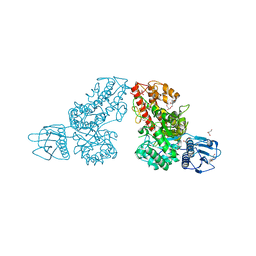 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with 4-O-methyl-glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K9D
 
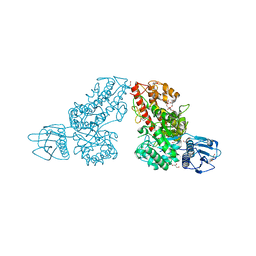 | | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1L8N
 
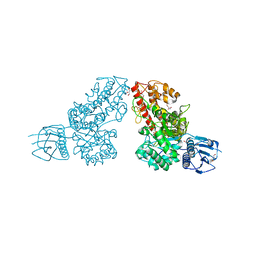 | | The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose | | Descriptor: | 4-O-methyl-beta-D-glucopyranuronic acid, ALPHA-D-GLUCURONIDASE, GLYCEROL, ... | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-03-21 | | Release date: | 2003-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1MQP
 
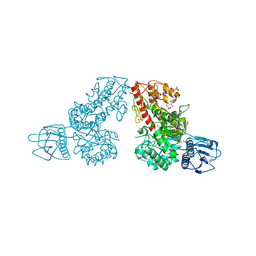 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1MQR
 
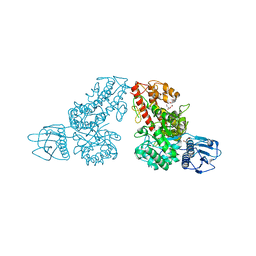 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE (E386Q) FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1MQQ
 
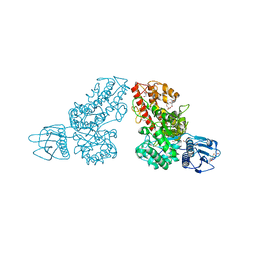 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL, alpha-D-glucopyranuronic acid | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
1K9F
 
 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
4A95
 
 | | Plasmodium vivax N-myristoyltransferase with quinoline inhibitor | | Descriptor: | 2-oxopentadecyl-CoA, 3-(3-BUTYL-6-METHOXY-2-METHYL-QUINOLIN-4-YL)SULFANYLPROPANENITRILE, CHLORIDE ION, ... | | Authors: | Goncalves, V, Brannigan, J.A, Whalley, D, Ansell, K.H, Saxty, B, Holder, A.A, Wilkinson, A.J, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2011-11-24 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Plasmodium Vivax N-Myristoyltransferase Inhibitors: Screening, Synthesis, and Structural Characterization of Their Binding Mode.
J.Med.Chem., 55, 2012
|
|
2WEY
 
 | | Human PDE-papaverine complex obtained by ligand soaking of cross- linked protein crystals | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Andersen, O.A, Schonfeld, D.L, Toogood-Johnson, I, Felicetti, B, Albrecht, C, Fryatt, T, Whittaker, M, Hallett, D, Barker, J. | | Deposit date: | 2009-04-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cross-Linking of Protein Crystals as an Aid in the Generation of Binary Protein-Ligand Crystal Complexes, Exemplified by the Human Pde10A-Papaverine Structure.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3S2O
 
 | | Fragment based discovery and optimisation of bace-1 inhibitors | | Descriptor: | (3S)-3-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-[(1R,3S,5R,7R)-tricyclo[3.3.1.1~3,7~]dec-2-yl]pentanamide, Beta-secretase 1, IODIDE ION | | Authors: | Madden, J, Godemann, R, Smith, M.A, Hallett, D, Barker, J, Kraemer, J. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4CNI
 
 | | Crystal structure of the Fab portion of Olokizumab in complex with IL- 6 | | Descriptor: | INTERLEUKIN-6, OLOKIZUMAB HEAVY CHAIN, FAB PORTION, ... | | Authors: | Shaw, S, Bourne, T, Meier, C, Carrington, B, Gelinas, R, Henry, A, Popplewell, A, Adams, R, Baker, T, Rapecki, S, Marshall, D, Neale, H, Lawson, A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-30 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Characterization of Olokizumab: A Humanized Antibody Targeting Interleukin-6 and Neutralizing Gp130-Signaling.
Mabs, 6, 2014
|
|
