4NEA
 
 | | 1.90 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NML
 
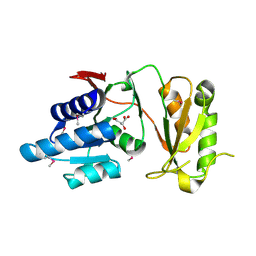 | | 2.60 Angstrom resolution crystal structure of putative ribose 5-phosphate isomerase from Toxoplasma gondii ME49 in complex with DL-Malic acid | | Descriptor: | CHLORIDE ION, D-MALATE, Ribulose 5-phosphate isomerase | | Authors: | Halavaty, A.S, Dubrovska, I, Flores, K, Shanmugam, D, Shuvalova, L, Roos, D, Ruan, J, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NLM
 
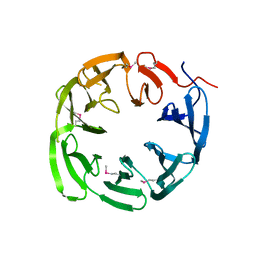 | | 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo1340 protein | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.18 Angstrom resolution crystal structure of uncharacterized protein lmo1340 from Listeria monocytogenes EGD-e
To be Published
|
|
4Q92
 
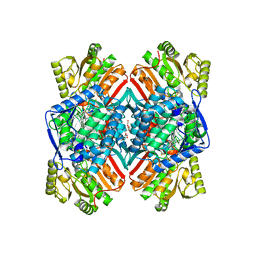 | | 1.90 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-modified Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NU9
 
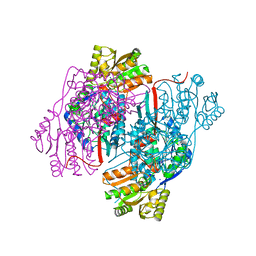 | | 2.30 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-free Cys289 | | Descriptor: | Betaine aldehyde dehydrogenase, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Stam, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QN2
 
 | | 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with NAD+ and BME-free Cys289 | | Descriptor: | ACETATE ION, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QJE
 
 | | 1.85 Angstrom resolution crystal structure of apo betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) with BME-free sulfinic acid form of Cys289 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QYI
 
 | | 1.95 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' with HEPES molecule in the active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' with HEPES molecule in the active site
To be Published
|
|
4QRI
 
 | | 2.35 Angstrom resolution crystal structure of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Flores, K.J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Leptospira interrogans serovar Copenhageni str. Fiocruz L1-130
To be Published
|
|
4QTO
 
 | | 1.65 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-modified Cys289 and PEG molecule in active site | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QUS
 
 | | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase YpeA, PHOSPHATE ION | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Flores, K.J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
To be Published
|
|
3NVT
 
 | | 1.95 Angstrom crystal structure of a bifunctional 3-deoxy-7-phosphoheptulonate synthase/chorismate mutase (aroA) from Listeria monocytogenes EGD-e | | Descriptor: | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, ACETATE ION, MANGANESE (II) ION | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of a 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain.
Protein Sci., 21, 2012
|
|
3PGJ
 
 | | 2.49 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | Authors: | Halavaty, A.S, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | 2.49 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate
To be Published
|
|
3OGA
 
 | | 1.75 Angstrom resolution crystal structure of a putative NTP pyrophosphohydrolase (yfaO) from Salmonella typhimurium LT2 | | Descriptor: | BETA-MERCAPTOETHANOL, Nucleoside triphosphatase nudI, PHOSPHATE ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of a putative NTP pyrophosphohydrolase (yfaO) from Salmonella typhimurium LT2
To be Published
|
|
3PAJ
 
 | | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | MAGNESIUM ION, Nicotinate-nucleotide pyrophosphorylase, carboxylating | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Shuvalova, L, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of a quinolinate phosphoribosyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3R38
 
 | | 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-15 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e
To be Published
|
|
3RYK
 
 | | 1.63 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase (rfbC) from Bacillus anthracis str. Ames with TDP and PPi bound | | Descriptor: | PYROPHOSPHATE 2-, THYMIDINE-5'-DIPHOSPHATE, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Halavaty, A.S, Kuhn, M, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-11 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose 3,5-epimerase (RfbC).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3RU6
 
 | | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, IODIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3OO2
 
 | | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL | | Descriptor: | Alanine racemase 1, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-30 | | Release date: | 2010-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | 2.37 Angstrom resolution crystal structure of an alanine racemase (alr) from Staphylococcus aureus subsp. aureus COL
TO BE PUBLISHED
|
|
3SC6
 
 | | 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, dTDP-4-dehydrorhamnose reductase | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-07 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose reductase (RfbD).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4D2J
 
 | | Crystal structure of F16BP Aldolase from Toxoplasma gondii (TgALD1) | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, FRUCTOSE-BISPHOSPHATE ALDOLASE, ... | | Authors: | Tonkin, M.L, Ramaswamy, R, Halavaty, A.S, Ruan, J, Igarashi, M, Ngo, H.M, Boulanger, M.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Functional Divergence of the Aldolase Fold in Toxoplasma Gondii.
J.Mol.Biol., 427, 2015
|
|
5DO8
 
 | | 1.8 Angstrom crystal structure of Listeria monocytogenes Lmo0184 alpha-1,6-glucosidase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Lmo0184 protein, ... | | Authors: | Light, S.H, Halavaty, A.S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-10 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure to function of an alpha-glucan metabolic pathway that promotes Listeria monocytogenes pathogenesis.
Nat Microbiol, 2, 2016
|
|
4RQ9
 
 | | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, GLYCEROL, ... | | Authors: | Woitowich, N.C, Halavaty, A.S, Gallagher, K.D, Nugent, A.C, Patel, H, Duong, P, Kovaleva, S.E, St.Peter, S, Ozarowski, W.B, Hernandez, C.N, Stojkovic, E.A. | | Deposit date: | 2014-10-31 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the chromophore-binding domain of Stigmatella aurantiaca bacteriophytochrome (Thr289His mutant) in the Pr state
To be Published
|
|
4O96
 
 | | 2.60 Angstrom resolution crystal structure of a protein kinase domain of type III effector NleH2 (ECs1814) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, type III effector protein kinase | | Authors: | Anderson, S.M, Halavaty, A.S, Wawrzak, Z, Kudritska, M, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Type III Effector NleH2 from Escherichia coli O157:H7 str. Sakai Features an Atypical Protein Kinase Domain.
Biochemistry, 53, 2014
|
|
4JBE
 
 | | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Filippova, E.V, Halavaty, A, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Gamma-glutamyl phosphate Reductase from Saccharomonospora viridis.
TO BE PUBLISHED
|
|
