7E9C
 
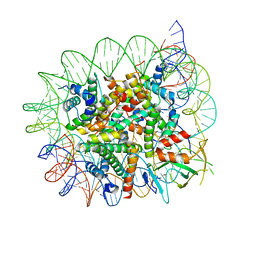 | | Cryo-EM structure of the 1:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
7E9F
 
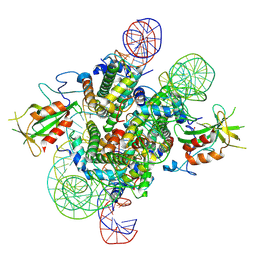 | | Cryo-EM structure of the 2:1 Orc1 BAH domain in complex with nucleosome | | Descriptor: | DNA (147-mer), Histone H2A.2, Histone H2B.2, ... | | Authors: | Jiang, H, Yu, C, Liu, C.P, Han, X, Yu, Z, Xu, R.M. | | Deposit date: | 2021-03-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Nucleosome binding relinquishes the association of the BAH domain of Orc1 with Sir1
To Be Published
|
|
6H6D
 
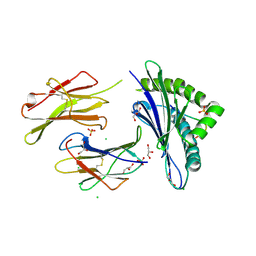 | |
6H6H
 
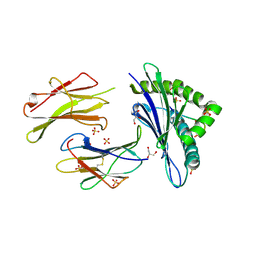 | |
8WDM
 
 | | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Li, Q, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis
To Be Published
|
|
8WDW
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium | | Descriptor: | GLYCEROL, SULFATE ION, UMG-SP2 | | Authors: | Cong, L, Li, Z.S, Gao, J, Li, Q, Chen, Y.Y, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2023-09-16 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium
To Be Published
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8AN4
 
 | | MenT1 toxin (rv0078a) from Mycobacterium tuberculosis H37Rv | | Descriptor: | Bacterial toxin | | Authors: | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
5M00
 
 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M01
 
 | | Crystal structure of murine P14 TCR/ H-2Db complex with PA, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
7C4J
 
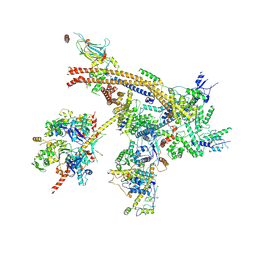 | | Cryo-EM structure of the yeast Swi/Snf complex in a nucleosome free state | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Regulator of Ty1 transposition protein 102, ... | | Authors: | Wang, C.C, Guo, Z.Y, Zhan, X.C, Zhang, X.F. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the yeast Swi/Snf complex in a nucleosome free state
Nat Commun, 2020
|
|
9IK1
 
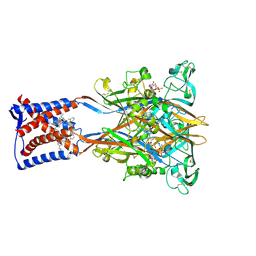 | | Cryo-EM structure of the human P2X3 receptor-compound 26a complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[2-cyclopropyl-7-[[(1~{R})-1-naphthalen-2-ylethyl]amino]-[1,2,4]triazolo[1,5-a]pyrimidin-5-yl]piperazine-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kim, S, Kim, G.R, Kim, Y.O, Han, X, Nagel, J, Kim, J, Song, D.I, Muller, C.E, Yoon, M.H, Jin, M.S, Kim, Y.C. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Discovery of Triazolopyrimidine Derivatives as Selective P2X3 Receptor Antagonists Binding to an Unprecedented Allosteric Site as Evidenced by Cryo-Electron Microscopy.
J.Med.Chem., 67, 2024
|
|
6U6V
 
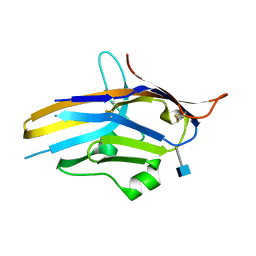 | | Crystal structure of human PD-1H / VISTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Slater, B.T, Han, X, Chen, L, Xiong, Y. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into T cell coinhibition by PD-1H (VISTA).
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8YSV
 
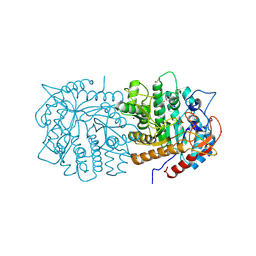 | | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase, ETHANOL, ... | | Authors: | Wang, W.Y, Li, Y.J, Liu, Z.Y, Han, X.D. | | Deposit date: | 2024-03-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis
To Be Published
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | Descriptor: | hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
8XTC
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, GLYCEROL, umgsp2-mut | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium in complex with ligand
To Be Published
|
|
8XTB
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, umgsp2 | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Wei, R, Liu, W.D, Uwe, B. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium
To Be Published
|
|
8XRZ
 
 | | Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis | | Descriptor: | 4-oxidanylbutyl ~{N}-[4-[(4-aminophenyl)methyl]phenyl]carbamate, Carboxylic ester hydrolase, SULFATE ION | | Authors: | Li, Z.S, Wang, H, Zheng, Z.R, Cong, L, Chen, Y.Y, Han, X, Wei, R, Uwe, B, liu, W.D. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis
To Be Published
|
|
7X63
 
 | | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex | | Descriptor: | BD-236 Fab heavy chain, BD-236 Fab light chain, Spike protein S1 | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-03-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | SARS-CoV-2-Beta-RBD and BD-236-GWP/P-VK antibody complex
To Be Published
|
|
7XIL
 
 | | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B38 Fab heavy chain, B38 Fab light chain, ... | | Authors: | Shi, R, Wang, Y, Wu, Z, Han, X, Yan, J. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | SARS-CoV-2-Beta-RBD and B38-GWP/P-VK antibody complex
To Be Published
|
|
7W66
 
 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W69
 
 | | Crystal structure of a PSH1 mutant in complex with EDO | | Descriptor: | 1,2-ETHANEDIOL, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
