8HSY
 
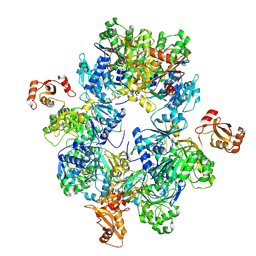 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | An inhibitory mechanism of AasS, an exogenous fatty acid scavenger: Implications for re-sensitization of FAS II antimicrobials.
Plos Pathog., 20, 2024
|
|
8I22
 
 | | Acyl-ACP synthetase structure bound to pimelic acid monoethyl ester | | Descriptor: | 7-ethoxy-7-oxidanylidene-heptanoic acid, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-17 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase
Nat.Struct.Mol.Biol., 2025
|
|
8I35
 
 | | Acyl-ACP synthetase structure bound to oleic acid | | Descriptor: | Acyl-acyl carrier protein synthetase, OLEIC ACID | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-16 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I3I
 
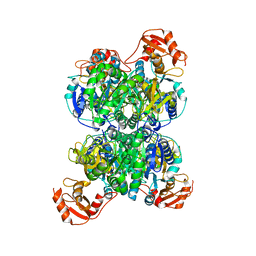 | | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2 | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8HZX
 
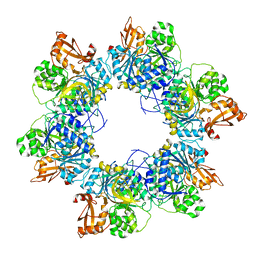 | | Acyl-ACP synthetase structure-2 | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I49
 
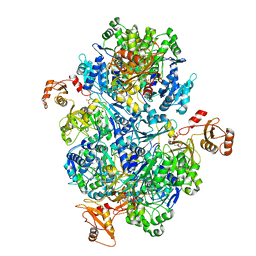 | | Acyl-ACP synthetase structure bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
1CX5
 
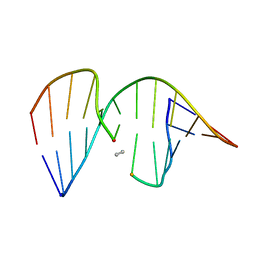 | | ANTISENSE DNA/RNA HYBRID CONTAINING MODIFIED BACKBONE | | Descriptor: | 5'-D(*CP*GP*CP*GP*TP*T*(MMT)P*TP*GP*CP*GP*C), 5'-R(*GP*CP*GP*CP*AP*AP*AP*AP*CP*GP*CP*G) | | Authors: | Yang, X, Han, X, Cross, C, Sanghvi, Y, Gao, X. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker.
Biochemistry, 38, 1999
|
|
6CDY
 
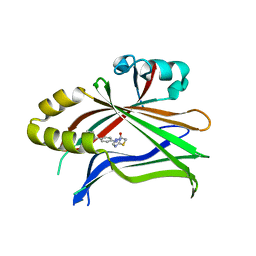 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
7WK0
 
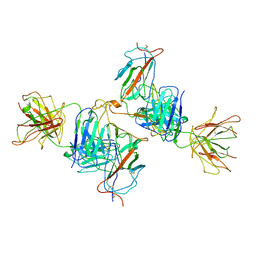 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
8I51
 
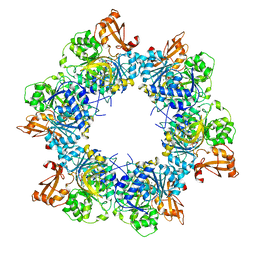 | | Acyl-ACP synthetase structure bound to AMP-MC7 | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-21 | | Release date: | 2024-01-24 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8I6M
 
 | | Acyl-ACP synthetase structure bound to AMP-C18:1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, MAGNESIUM ION, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I8D
 
 | | Acyl-ACP synthetase structure bound to MC7-ACP | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
8I8E
 
 | | Acyl-ACP synthetase structure bound to C18:1-ACP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl carrier protein, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structure and catalytic mechanism of exogenous fatty acid recycling by AasS, a versatile acyl-ACP synthetase.
Nat.Struct.Mol.Biol., 2025
|
|
5XNO
 
 | | Structure of M-LHCII and CP24 complexes in the unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNL
 
 | | Structure of stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7WHJ
 
 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHK
 
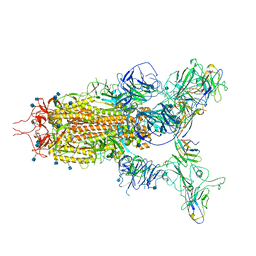 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHI
 
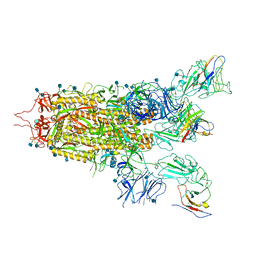 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
