6LOM
 
 | |
3TC1
 
 | | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori | | 分子名称: | MAGNESIUM ION, Octaprenyl Pyrophosphate Synthase | | 著者 | Zhang, J.Y, Zhang, X.L, Li, D.F, Zou, Q.M, Wang, D.C. | | 登録日 | 2011-08-08 | | 公開日 | 2011-08-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Octaprenyl Pyrophosphate Synthase from Helicobacter pylori
To be Published
|
|
4N2B
 
 | | Crystal structure of Protein Arginine Deiminase 2 (10 mM Ca2+) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | 著者 | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | 登録日 | 2013-10-04 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
1FZX
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAAAAACGG | | 分子名称: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*AP*AP*CP*GP*G)-3' | | 著者 | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | 登録日 | 2000-10-04 | | 公開日 | 2001-03-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
1EUE
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | 分子名称: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Oganesyan, V, Zhang, X. | | 登録日 | 2000-04-19 | | 公開日 | 2001-04-04 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Modulation of redox potential in electron transfer proteins: effects of complex formation on the active site microenvironment of cytochrome b5.
FARADAY DISC.CHEM.SOC, 116, 2001
|
|
4R2E
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | 著者 | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | 登録日 | 2014-08-11 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
1G14
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAGAAACGG | | 分子名称: | 5'-D(*CP*CP*GP*TP*TP*TP*CP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*GP*AP*AP*AP*CP*GP*G)-3' | | 著者 | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | 登録日 | 2000-10-10 | | 公開日 | 2001-03-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
2Q13
 
 | |
1BR0
 
 | | THREE DIMENSIONAL STRUCTURE OF THE N-TERMINAL DOMAIN OF SYNTAXIN 1A | | 分子名称: | PROTEIN (SYNTAXIN 1-A) | | 著者 | Fernandez, I, Ubach, J, Dubulova, I, Zhang, X, Sudhof, T.C, Rizo, J. | | 登録日 | 1998-08-25 | | 公開日 | 1998-09-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A.
Cell(Cambridge,Mass.), 94, 1998
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | 分子名称: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | 分子名称: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | 著者 | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | 登録日 | 2018-09-21 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IZK
 
 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | 分子名称: | CHLORIDE ION, IMIDAZOLE, L(+)-TARTARIC ACID, ... | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2018-12-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus
To Be Published
|
|
2L37
 
 | | 3D solution structure of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical) | | 分子名称: | Ribosome-inactivating protein luffin P1 | | 著者 | Ng, Y.M, Yang, Y, Sze, K.H, Zhang, X, Zheng, Y.T, Shaw, P.C. | | 登録日 | 2010-09-08 | | 公開日 | 2011-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical).
J.Struct.Biol., 2010
|
|
2LUH
 
 | | NMR structure of the Vta1-Vps60 complex | | 分子名称: | Vacuolar protein sorting-associated protein VTA1, Vacuolar protein-sorting-associated protein 60 | | 著者 | Yang, Z, Vild, C, Ju, J, Zhang, X, Liu, J, Shen, J, Zhao, B, Lan, W, Gong, F, Liu, M, Cao, C, Xu, Z. | | 登録日 | 2012-06-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of Molecular Recognition between ESCRT-III-like Protein Vps60 and AAA-ATPase Regulator Vta1 in the Multivesicular Body Pathway.
J.Biol.Chem., 287, 2012
|
|
7X2U
 
 | | Structure of a human NHE3-CHP1 complex in the autoinhibited state | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 3, ... | | 著者 | Dong, Y, Li, H, Gao, Y, Zhang, X.C, Zhao, Y. | | 登録日 | 2022-02-26 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of a human NHE3-CHP1 complex in the autoinhibited state
Sci Adv, 2022
|
|
3T9N
 
 | | Crystal structure of a membrane protein | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Small-conductance mechanosensitive channel | | 著者 | Yang, M, Zhang, X, Ge, J, Wang, J. | | 登録日 | 2011-08-03 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.456 Å) | | 主引用文献 | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8W9C
 
 | |
4IP3
 
 | | Complex structure of OspI and Ubc13 | | 分子名称: | ORF169b, Ubiquitin-conjugating enzyme E2 N | | 著者 | Fu, P, Jin, M, Zhang, X, Xu, L, Xia, Z, Zhu, Y. | | 登録日 | 2013-01-09 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure Analysis of Ubc13 Inactivation
To be Published
|
|
3P2D
 
 | |
4JGC
 
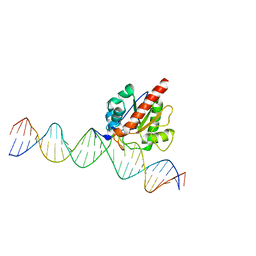 | | Human TDG N140A mutant IN A COMPLEX WITH 5-carboxylcytosine (5caC) | | 分子名称: | 4-amino-2-oxo-1,2-dihydropyrimidine-5-carboxylic acid, G/T mismatch-specific thymine DNA glycosylase, oligonucleotide, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2013-02-28 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.582 Å) | | 主引用文献 | Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Dna Repair, 12, 2013
|
|
7X5A
 
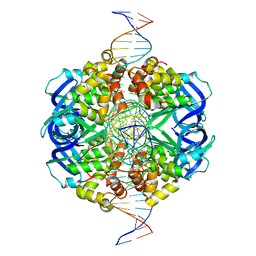 | | CryoEM structure of RuvA-Holliday junction complex | | 分子名称: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-04 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
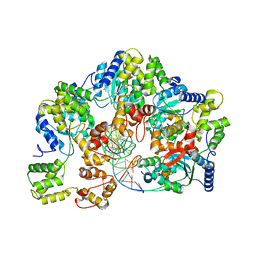 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | 分子名称: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
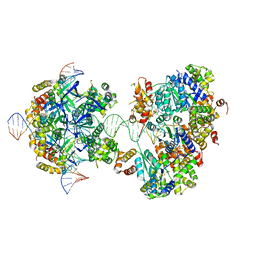 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | 分子名称: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | 著者 | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | 登録日 | 2022-03-10 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | ELECTRON MICROSCOPY (7.02 Å) | | 主引用文献 | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | 分子名称: | Minor spike protein H | | 著者 | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | 登録日 | 2013-03-19 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | 分子名称: | Protein kinase family protein | | 著者 | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | 登録日 | 2019-11-13 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
