6Q2O
 
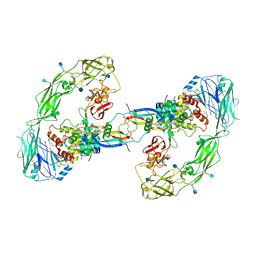 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | 著者 | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | 登録日 | 2019-08-08 | | 公開日 | 2019-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
1AQL
 
 | |
190D
 
 | |
2M9M
 
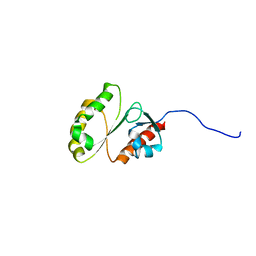 | | Solution Structure of ERCC4 domain of human FAAP24 | | 分子名称: | Fanconi anemia-associated protein of 24 kDa | | 著者 | Wu, F, Han, X, Shi, C, Gong, W, Tian, C. | | 登録日 | 2013-06-18 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure analysis of FAAP24 reveals single-stranded DNA-binding activity and domain functions in DNA damage response.
Cell Res., 23, 2013
|
|
2M9N
 
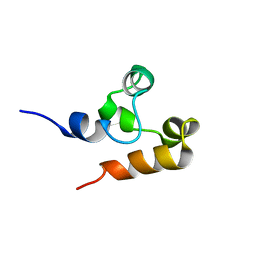 | | Solution Structure of (HhH)2 domain of human FAAP24 | | 分子名称: | Fanconi anemia-associated protein of 24 kDa | | 著者 | Wu, F, Han, X, Shi, C, Gong, W, Tian, C. | | 登録日 | 2013-06-18 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure analysis of FAAP24 reveals single-stranded DNA-binding activity and domain functions in DNA damage response.
Cell Res., 23, 2013
|
|
1AWP
 
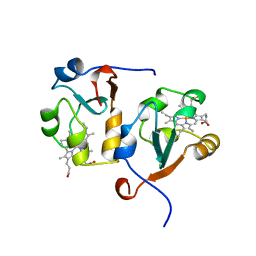 | |
6Q2N
 
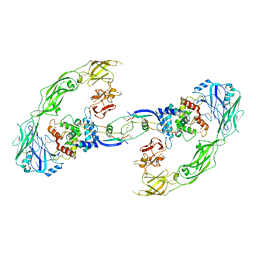 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | 分子名称: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | 著者 | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | 登録日 | 2019-08-08 | | 公開日 | 2019-10-02 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
173L
 
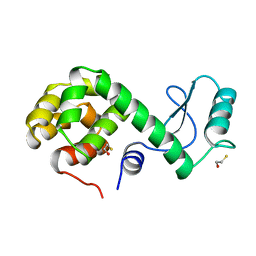 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | 著者 | Xiong, X.-P, Zhang, X.-J, Sun, D, Matthews, B.W. | | 登録日 | 1995-03-24 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
7OZR
 
 | |
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | 分子名称: | NADP-dependent isopropanol dehydrogenase | | 著者 | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | 登録日 | 2022-04-21 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
1B5M
 
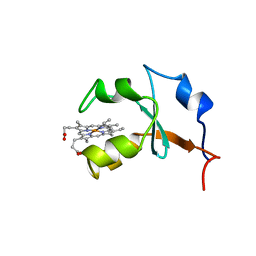 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | 分子名称: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Rivera, M, White, S.P, Zhang, X. | | 登録日 | 1996-11-07 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | 13C NMR spectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c.
Biochemistry, 35, 1996
|
|
7VX2
 
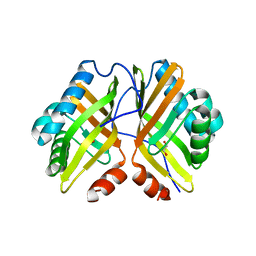 | | Crystal Structure of the Y53F/N55A/I80F/L114V/I116V mutant of LEH | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | 登録日 | 2021-11-12 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.485 Å) | | 主引用文献 | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWM
 
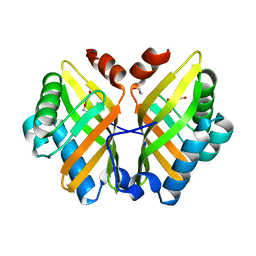 | | Crystal Structure of the Y53F/N55A/I116V mutant of LEH | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | 登録日 | 2021-11-11 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
7VWD
 
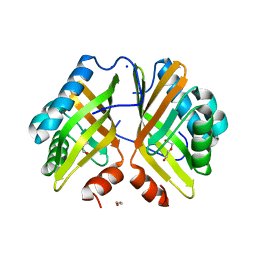 | | Crystal Structure of the Y53F/N55A mutant of LEH | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | 登録日 | 2021-11-10 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Rational enzyme design for enabling biocatalytic Baldwin cyclization and asymmetric synthesis of chiral heterocycles.
Nat Commun, 13, 2022
|
|
177L
 
 | |
172L
 
 | |
7CG7
 
 | | Cryo-EM structure of the flagellar MS ring with C34 symmetry from Salmonella | | 分子名称: | Flagellar M-ring protein | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.61 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CBL
 
 | | Cryo-EM structure of the flagellar LP ring from Salmonella | | 分子名称: | Flagellar L-ring protein, Flagellar P-ring protein, OCTANOIC ACID (CAPRYLIC ACID) | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-12 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-05-26 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGB
 
 | | Cryo-EM structure of the flagellar hook from Salmonella | | 分子名称: | Flagellar hook protein FlgE | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-07-01 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGO
 
 | | Cryo-EM structure of the flagellar motor-hook complex from Salmonella | | 分子名称: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar MS ring L1, ... | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-07-01 | | 公開日 | 2021-04-28 | | 最終更新日 | 2021-05-26 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG4
 
 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | 分子名称: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CBM
 
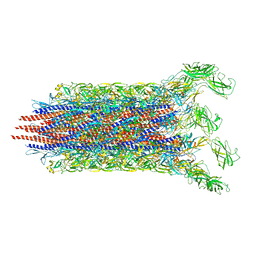 | | Cryo-EM structure of the flagellar distal rod with partial hook from Salmonella | | 分子名称: | Flagellar basal-body rod protein FlgF, Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-12 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG0
 
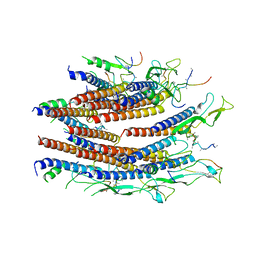 | | Cryo-EM structure of the flagellar proximal rod with FliF peptides from Salmonella | | 分子名称: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | 著者 | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
167L
 
 | |
180L
 
 | |
