3QXL
 
 | | Crystal structure of the CDC25 Domain from Ral-specific Guanine-nucleotide Exchange Factor RalGPS1a | | Descriptor: | Ras-specific guanine nucleotide-releasing factor RalGPS1 | | Authors: | Peng, W, Xu, J, Guan, X, Sun, Y, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural study of the Cdc25 domain from Ral-specific guanine-nucleotide exchange factor RalGPS1a.
Protein Cell, 2, 2011
|
|
1G14
 
 | | NMR SOLUTION STRUCTURE OF THE DNA DODECAMER GGCAAGAAACGG | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*CP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*GP*AP*AP*AP*CP*GP*G)-3' | | Authors: | MacDonald, D, Herbert, K, Zhang, X, Pologruto, T, Lu, P. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an A-tract DNA bend.
J.Mol.Biol., 306, 2001
|
|
2Z41
 
 | | Crystal Structure Analysis of the Ski2-type RNA helicase | | Descriptor: | MAGNESIUM ION, putative ski2-type helicase | | Authors: | Nakashima, T, Zhang, X, Kakuta, Y, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3
Protein Sci., 17, 2008
|
|
3EA2
 
 | | Crystal Structure of the Myo-inositol bound Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
1TU3
 
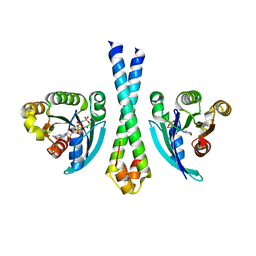 | | Crystal Structure of Rab5 complex with Rabaptin5 C-terminal Domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab GTPase binding effector protein 1, ... | | Authors: | Zhu, G, Zhai, P, Liu, J, Terzyan, S, Li, G, Zhang, X.C. | | Deposit date: | 2004-06-24 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of Rab5-Rabaptin5 interaction in endocytosis
Nat.Struct.Mol.Biol., 11, 2004
|
|
8GWA
 
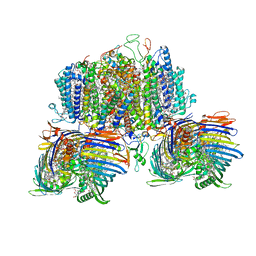 | | Structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2022-09-16 | | Release date: | 2022-11-23 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-electron microscopy structure of the intact photosynthetic light-harvesting antenna-reaction center complex from a green sulfur bacterium.
J Integr Plant Biol, 65, 2023
|
|
4EJZ
 
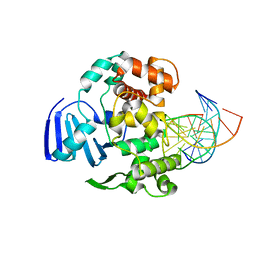 | | Structure of MBOgg1 in complex with low affinity DNA ligand | | Descriptor: | 3-Methyladenine DNA glycosylase, DNA (5'-D(*AP*GP*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*T*GP*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3') | | Authors: | Jiang, T, Yu, H.J, Bi, L.J, Zhang, X.E, Yang, M.Z. | | Deposit date: | 2012-04-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of MBOgg1 in complex with two abasic DNA ligands
J.Struct.Biol., 181, 2013
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XEX
 
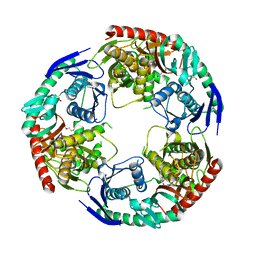 | | Crystal structure of S.aureus PNPase catalytic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYROPHOSPHATE, ... | | Authors: | Wang, X, Zhang, X, Zang, J. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enolase binds to RnpA in competition with PNPase in Staphylococcus aureus
FEBS Lett., 591, 2017
|
|
8HI2
 
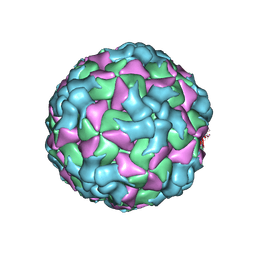 | |
8HHS
 
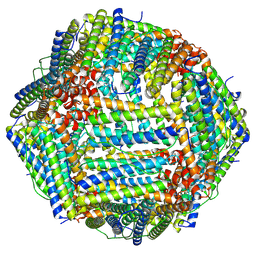 | |
1QQR
 
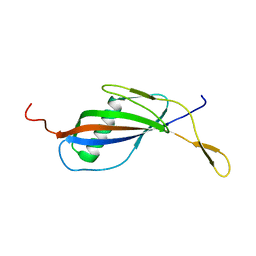 | | CRYSTAL STRUCTURE OF STREPTOKINASE DOMAIN B | | Descriptor: | STREPTOKINASE DOMAIN B | | Authors: | Spraggon, G, Zhang, X.X, Ponting, C.P, Fox, V.F, Phillips, C, Smith, R.A.G, Jones, E.Y, Dobson, C, Stuart, D.I. | | Deposit date: | 1999-06-07 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptokinse Domain B
To be Published
|
|
6LT0
 
 | | cryo-EM structure of C9ORF72-SMCR8-WDR41 | | Descriptor: | Guanine nucleotide exchange C9orf72, Guanine nucleotide exchange protein SMCR8, WD repeat-containing protein 41 | | Authors: | Tang, D, Sheng, J, Xu, L, Zhan, X, Yan, C, Qi, S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7WB4
 
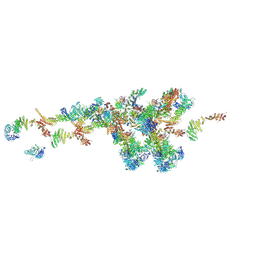 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
3F8I
 
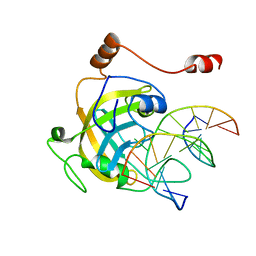 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
223L
 
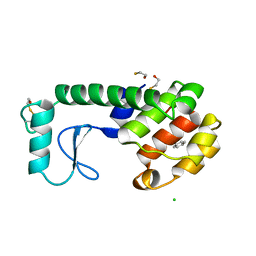 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
3KV9
 
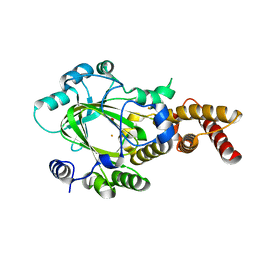 | | Structure of KIAA1718 Jumonji domain | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KXL
 
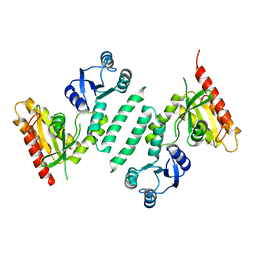 | | crystal structure of SsGBP mutation variant G235S | | Descriptor: | GTP-binding protein (HflX), THIOCYANATE ION | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
222L
 
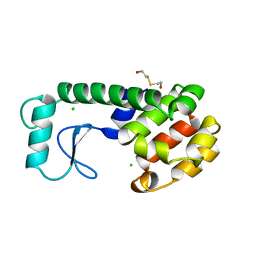 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
5X5S
 
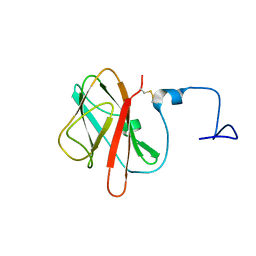 | |
3WY7
 
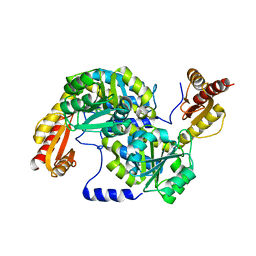 | | Crystal structure of Mycobacterium smegmatis 7-Keto-8-aminopelargonic acid (KAPA) synthase BioF | | Descriptor: | 8-amino-7-oxononanoate synthase | | Authors: | Fan, S.H, Li, D.F, Wang, D.C, Chen, G.J, Zhang, X.E, Bi, L.J. | | Deposit date: | 2014-08-20 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Mycobacterium smegmatis 7-keto-8-aminopelargonic acid (KAPA) synthase
Int.J.Biochem.Cell Biol., 58C, 2014
|
|
3KXI
 
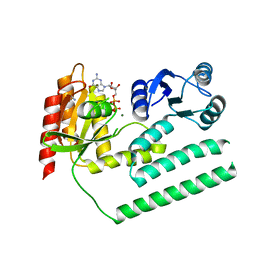 | | crystal structure of SsGBP and GDP complex | | Descriptor: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
227L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
