1IAQ
 
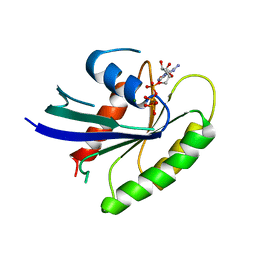 | | C-H-RAS P21 PROTEIN MUTANT WITH THR 35 REPLACED BY SER (T35S) COMPLEXED WITH GUANOSINE-5'-[B,G-IMIDO] TRIPHOSPHATE | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Spoerner, M, Herrmann, C, Vetter, I.R, Kalbitzer, H.R, Wittinghofer, A. | | Deposit date: | 2001-03-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic properties of the Ras switch I region and its importance for binding to effectors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GUW
 
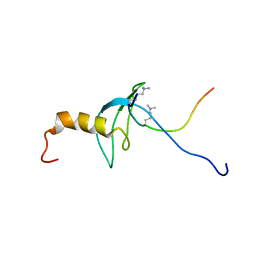 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
1HXN
 
 | |
7QIG
 
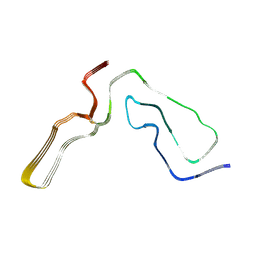 | | Infectious mouse-adapted RML scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Zhang, W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2021-12-14 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | 2.7 angstrom cryo-EM structure of ex vivo RML prion fibrils.
Nat Commun, 13, 2022
|
|
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
8EJJ
 
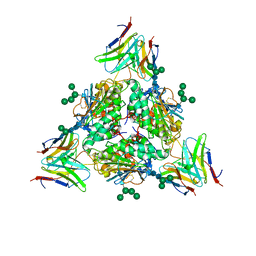 | | Lassa virus glycoprotein complex (Josiah) bound to S370.7 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJE
 
 | | Structure of lineage II Lassa virus glycoprotein complex (strain NIG08-A41) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein GP1, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJF
 
 | | Structure of lineage V Lassa virus glycoprotein complex (strain Soromba-R) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJH
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 12.1F Fab | | Descriptor: | 12.1F Fab heavy chain, 12.1F Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJI
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab | | Descriptor: | 19.7E Fab heavy chain, 19.7E Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJD
 
 | | Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJG
 
 | |
7QSM
 
 | | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSN
 
 | | Bovine complex I in lipid nanodisc, Deactive-apo | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSL
 
 | | Bovine complex I in lipid nanodisc, Active-apo | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSK
 
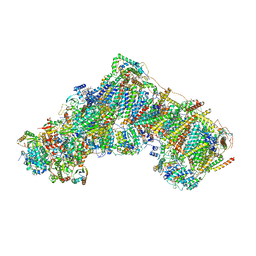 | | Bovine complex I in lipid nanodisc, Active-Q10 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSO
 
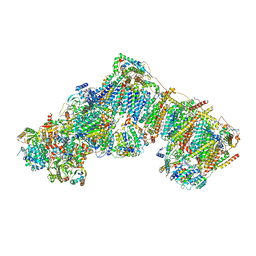 | | Bovine complex I in lipid nanodisc, State 3 (Slack) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QZD
 
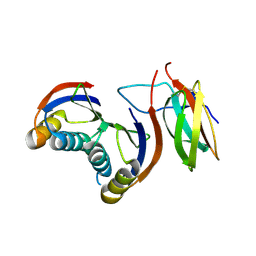 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikF with an engineered HMA domain of Pikp-1 (Pikp-SNK-EKE) from rice (Oryza sativa) | | Descriptor: | Avr-Pik, Resistance protein Pikp-1 | | Authors: | Maidment, J.H.R, Franceschetti, M, Longya, A, Banfield, M.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor.
Elife, 12, 2023
|
|
7QPX
 
 | |
1FMH
 
 | |
1FAQ
 
 | |
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
1GLB
 
 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, ... | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
1GLF
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-10-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1GLA
 
 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, GLYCEROL KINASE | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
