1I35
 
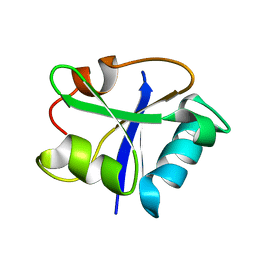 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF THE PROTEIN KINASE BYR2 FROM SCHIZOSACCHAROMYCES POMBE | | 分子名称: | PROTEIN KINASE BYR2 | | 著者 | Gronwald, W, Huber, F, Grunewald, P, Sporner, M, Wohlgemuth, S, Herrmann, C, Kalbitzer, H.R. | | 登録日 | 2001-02-13 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Ras binding domain of the protein kinase Byr2 from Schizosaccharomyces pombe.
Structure, 9, 2001
|
|
4JUD
 
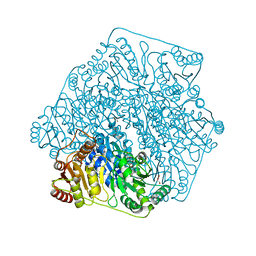 | |
1QJL
 
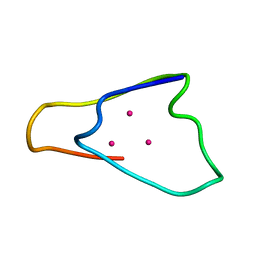 | | METALLOTHIONEIN MTA FROM SEA URCHIN (BETA DOMAIN) | | 分子名称: | CADMIUM ION, METALLOTHIONEIN | | 著者 | Riek, R, Precheur, B, Wang, Y, Mackay, E.A, Wider, G, Guntert, P, Liu, A, Kaegi, J.H.R, Wuthrich, K. | | 登録日 | 1999-06-24 | | 公開日 | 1999-08-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the sea urchin (Strongylocentrotus purpuratus) metallothionein MTA.
J. Mol. Biol., 291, 1999
|
|
4JUC
 
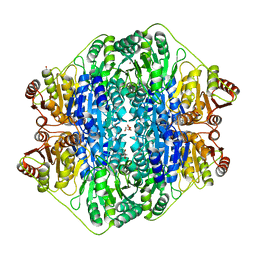 | |
1H1H
 
 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | 分子名称: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | 著者 | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | 登録日 | 2002-07-15 | | 公開日 | 2002-10-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
4JUA
 
 | |
1SGH
 
 | | Moesin FERM domain bound to EBP50 C-terminal peptide | | 分子名称: | Ezrin-radixin-moesin binding phosphoprotein 50, Moesin | | 著者 | Finnerty, C.M, Chambers, D, Ingraffea, J, Faber, H.R, Karplus, P.A, Bretscher, A. | | 登録日 | 2004-02-23 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain
J.Cell.Sci., 117, 2004
|
|
1QFR
 
 | | NMR SOLUTION STRUCTURE OF PHOSPHOCARRIER PROTEIN HPR FROM ENTEROCOCCUS FAECALIS | | 分子名称: | PHOSPHOCARRIER PROTEIN HPR | | 著者 | Maurer, T, Doeker, R, Goerler, A, Hengstenberg, W, Kalbitzer, H.R. | | 登録日 | 1999-04-13 | | 公開日 | 2001-02-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution.
Eur.J.Biochem., 268, 2001
|
|
4K9M
 
 | |
4K9P
 
 | |
4K9O
 
 | |
1HF0
 
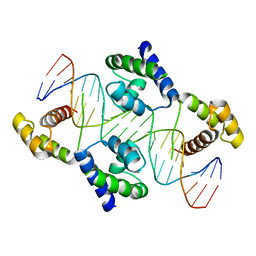 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | 分子名称: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | 著者 | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | 登録日 | 2000-11-27 | | 公開日 | 2001-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
4K9L
 
 | |
1QA5
 
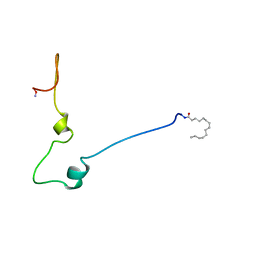 | |
1PW8
 
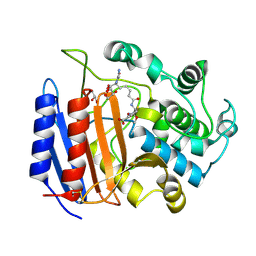 | | Covalent Acyl Enzyme Complex Of The R61 DD-Peptidase with A Highly Specific Cephalosporin | | 分子名称: | (6R,7R)-3-[(ACETYLOXY)METHYL]-7-{[(6S)-6-(GLYCYLAMINO)-7-OXIDO-7-OXOHEPTANOYL]AMINO}-8-OXO-5-THIA-1-AZABICYCLO[4.2.0]OCTANE-2-CARBOXYLATE, D-alanyl-D-alanine carboxypeptidase, GLYCEROL | | 著者 | Silvaggi, N.R, Josephine, H.R, Pratt, R.F, Kelly, J.A. | | 登録日 | 2003-07-01 | | 公開日 | 2004-07-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal structures of complexes between the R61 DD-peptidase and peptidoglycan-mimetic beta-lactams: a non-covalent complex with a "perfect penicillin"
J.Mol.Biol., 345, 2005
|
|
1PV7
 
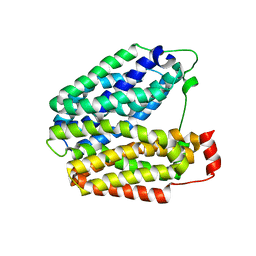 | | Crystal structure of lactose permease with TDG | | 分子名称: | Lactose permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | 著者 | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | 登録日 | 2003-06-26 | | 公開日 | 2003-08-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
1GRU
 
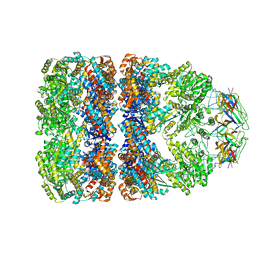 | | SOLUTION STRUCTURE OF GROES-ADP7-GROEL-ATP7 COMPLEX BY CRYO-EM | | 分子名称: | GROEL, GROES | | 著者 | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | 登録日 | 2001-12-16 | | 公開日 | 2002-01-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (12.5 Å) | | 主引用文献 | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1GR5
 
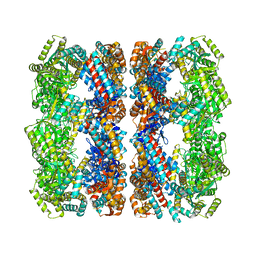 | | Solution Structure of apo GroEL by Cryo-Electron microscopy | | 分子名称: | 60 KDA CHAPERONIN | | 著者 | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | 登録日 | 2001-12-14 | | 公開日 | 2002-01-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.9 Å) | | 主引用文献 | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
4JU8
 
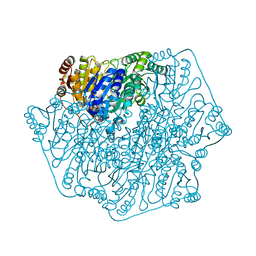 | |
1HK6
 
 | | Ral binding domain from Sec5 | | 分子名称: | EXOCYST COMPLEX COMPONENT SEC5 | | 著者 | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | 登録日 | 2003-03-05 | | 公開日 | 2003-03-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
4K9K
 
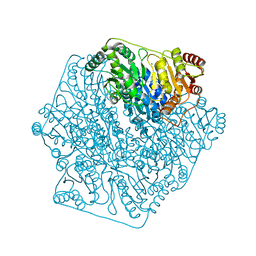 | |
1S4Z
 
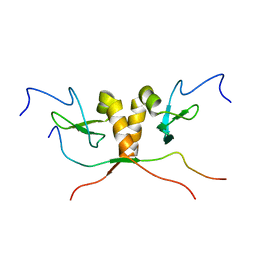 | | HP1 chromo shadow domain in complex with PXVXL motif of CAF-1 | | 分子名称: | Chromatin assembly factor 1 subunit A, Chromobox protein homolog 1 | | 著者 | Thiru, A, Nietlispach, D, Mott, H.R, Okuwaki, M, Lyon, D, Nielsen, P.R, Hirshberg, M, Verreault, A, Murzina, N.V, Laue, E.D. | | 登録日 | 2004-01-19 | | 公開日 | 2004-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin.
Embo J., 23, 2004
|
|
1GUW
 
 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | 分子名称: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | 著者 | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | 登録日 | 2002-02-01 | | 公開日 | 2002-03-12 | | 最終更新日 | 2018-01-17 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
4JUF
 
 | |
4JU9
 
 | |
