8CA3
 
 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA1
 
 | | Cryo-EM structure of the ACADVL dimer from Mus musculus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8EJI
 
 | | Lassa virus glycoprotein complex (Josiah) bound to 19.7E Fab | | Descriptor: | 19.7E Fab heavy chain, 19.7E Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
8EJG
 
 | |
8EJD
 
 | | Structure of lineage IV Lassa virus glycoprotein complex (strain Josiah) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Perrett, H.R, Ward, A.B. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural conservation of Lassa virus glycoproteins and recognition by neutralizing antibodies.
Cell Rep, 42, 2023
|
|
4A8A
 
 | | Asymmetric cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozyme | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14.2 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AAU
 
 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4BGC
 
 | |
4AB2
 
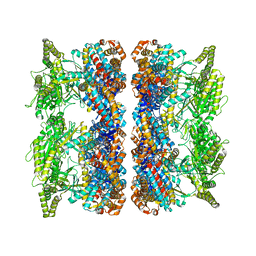 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4A8B
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 12-mer in complex with lysozymes | | Descriptor: | LYSOZYME C, PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-20 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AAS
 
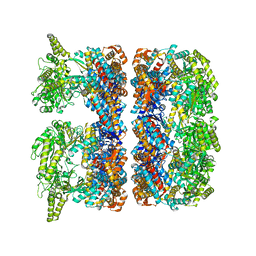 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4BRZ
 
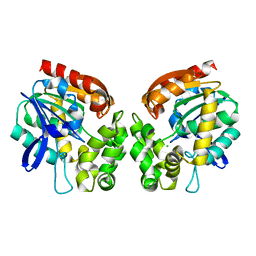 | | Haloalkane dehalogenase | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-06-06 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
4CF5
 
 | |
4CE6
 
 | |
4C6H
 
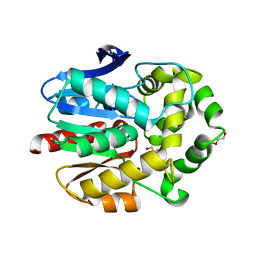 | | Haloalkane dehalogenase with 1-hexanol | | Descriptor: | CHLORIDE ION, HALOALKANE DEHALOGENASE, HEXAN-1-OL, ... | | Authors: | Novak, H.R, Sayer, C, Isupov, M, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Biochemical and Structural Characterisation of a Haloalkane Dehalogenase from a Marine Rhodobacteraceae.
FEBS Lett., 588, 2014
|
|
2BNA
 
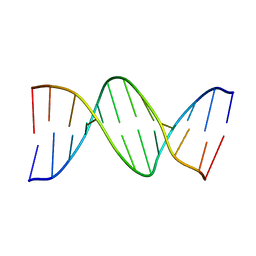 | |
2C5I
 
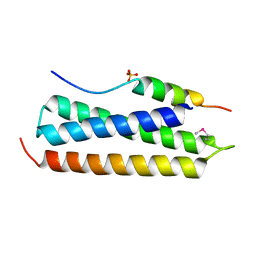 | | N-terminal domain of tlg1 complexed with N-terminus of vps51 in distorted conformation | | Descriptor: | SULFATE ION, T-SNARE AFFECTING A LATE GOLGI COMPARTMENT PROTEIN 1, VACUOLAR PROTEIN SORTING PROTEIN 51 | | Authors: | Fridmann-Sirkis, Y, Kent, H.M, Lewis, M.J, Evans, P.R, Pelham, H.R.B. | | Deposit date: | 2005-10-27 | | Release date: | 2006-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Interaction between the Snare Tlg1 and Vps51.
Traffic, 7, 2006
|
|
2C6F
 
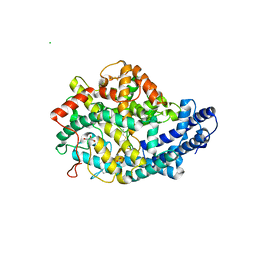 | | Structure of human somatic angiontensin-I converting enzyme N domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Corradi, H.R, Schwager, S.L.U, Nichinda, A, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of the N Domain of Human Somatic Angiotensin I-Converting Enzyme Provides a Structural Basis for Domain-Specific Inhibitor Design.
J.Mol.Biol., 357, 2006
|
|
4CF3
 
 | |
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BYU
 
 | | Negative stain EM reconstruction of M.tuberculosis Acr1(Hsp 16.3) fitted with wheat sHSP dimer | | Descriptor: | HEAT SHOCK PROTEIN 16.9B | | Authors: | Kennaway, C.K, Benesch, J.L.P, Gohlke, U, Wang, L, Robinson, C.V, Orlova, E.V, Saibil, H.R, Keep, N.H. | | Deposit date: | 2005-08-05 | | Release date: | 2005-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dodecameric Structure of the Small Heat Shock Protein Acr1 from Mycobacterium Tuberculosis.
J.Biol.Chem., 280, 2005
|
|
4A9G
 
 | | Symmetrized cryo-EM reconstruction of E. coli DegQ 24-mer in complex with beta-casein | | Descriptor: | PERIPLASMIC PH-DEPENDENT SERINE ENDOPROTEASE DEGQ | | Authors: | Malet, H, Canellas, F, Sawa, J, Yan, J, Thalassinos, K, Ehrmann, M, Clausen, T, Saibil, H.R. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Newly Folded Substrates Inside the Molecular Cage of the Htra Chaperone Degq
Nat.Struct.Mol.Biol., 19, 2012
|
|
2C5K
 
 | | N-terminal domain of tlg1 complexed with N-terminus of vps51 | | Descriptor: | SULFATE ION, T-SNARE AFFECTING A LATE GOLGI COMPARTMENT PROTEIN 1, VACUOLAR PROTEIN SORTING PROTEIN 51 | | Authors: | Fridmann-Sirkis, Y, Kent, H.M, Lewis, M.J, Evans, P.R, Pelham, H.R.B. | | Deposit date: | 2005-10-27 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of the Interaction between the Snare Tlg1 and Vps51.
Traffic, 7, 2006
|
|
2BFR
 
 | | The Macro domain is an ADP-ribose binding module | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HYPOTHETICAL PROTEIN AF1521, MAGNESIUM ION | | Authors: | Karras, G.I, Buhecha, H.R, Allen, M.D, Pugieux, C, Sait, F, Bycroft, M, Ladurner, A.G. | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Macro Domain is an Adp-Ribose Binding Module.
Embo J., 24, 2005
|
|
