2M61
 
 | | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails | | Descriptor: | Conotoxin Ar1446 | | Authors: | Sarma, S.P, Rajesh, R.P, Kumar, G.S, Sudarslal, S, Sabareesh, V, Gowd, K.H, Gupta, K, Krishnan, K.S, Balaram, P. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-16 | | Method: | SOLUTION NMR | | Cite: | NMR and Mass Spectrometric Studies of M-2 Branch Mini-M Conotoxins from Indian Cone Snails
To be Published
|
|
2LEH
 
 | | Solution structure of the core SMN-Gemin2 complex | | Descriptor: | Survival motor neuron protein, Survival of motor neuron protein-interacting protein 1 | | Authors: | Sarachan, K.L, Valentine, K, Gupta, K, Moorman, V, Gledhill, J, Bernens, M, Tommos, C, Wand, A.J, Van Duyne, G. | | Deposit date: | 2011-06-15 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the core SMN-Gemin2 complex.
Biochem.J., 445, 2012
|
|
4OT9
 
 | | crystal structure of the C-terminal domain of p100/NF-kB2 | | Descriptor: | Nuclear factor NF-kappa-B p100 subunit, SULFATE ION | | Authors: | Tao, Z.H, Huang, D.B, Fusco, A, Gupta, K, Ware, C.F, Duynne, G.V. | | Deposit date: | 2014-02-13 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | p100/I kappa B delta sequesters and inhibits NF-kappa B through kappaBsome formation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1HT5
 
 | | THE 2.75 ANGSTROM RESOLUTION MODEL OF OVINE COX-1 COMPLEXED WITH METHYL ESTER FLURBIPROFEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN METHYL ESTER, PROSTAGLANDIN H2 SYNTHASE-1, ... | | Authors: | Selinsky, B.S, Gupta, K, Sharkey, C.T, Loll, P.J. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
7OD3
 
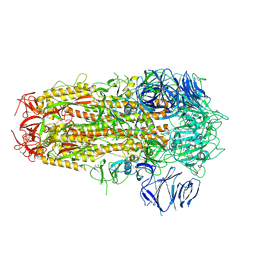 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ODL
 
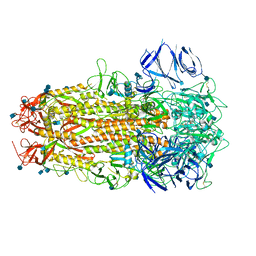 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
1P4W
 
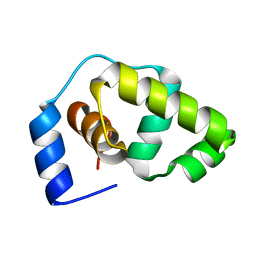 | | Solution structure of the DNA-binding domain of the Erwinia amylovora RcsB protein | | Descriptor: | rcsB | | Authors: | Pristovsek, P, Sengupta, K, Loehr, F, Schaefer, B, Wehland von Trebra, M, Rueterjans, H, Bernhard, F. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box.
J.Biol.Chem., 278, 2003
|
|
