7E4H
 
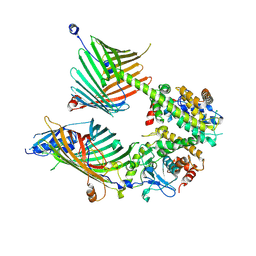 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7D3Y
 
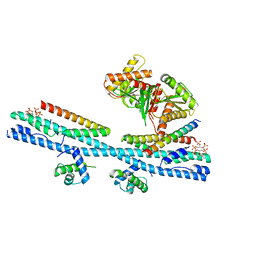 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
7V9U
 
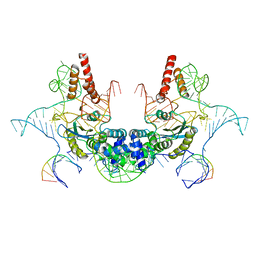 | | Cryo-EM structure of E.coli retron-Ec86 (RT-msDNA-RNA) at 3.2 angstrom | | Descriptor: | DNA (105-MER), RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*G)-3'), RNA (81-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2021-08-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
7VGG
 
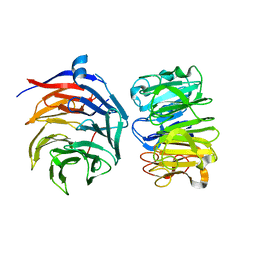 | | Cryo-EM structure of Ultraviolet-B activated UVR8 in complex with COP1 | | Descriptor: | E3 ubiquitin-protein ligase COP1, Ultraviolet-B receptor UVR8 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, Yin, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into UV-B-activated UVR8 bound to COP1.
Sci Adv, 8, 2022
|
|
7XJG
 
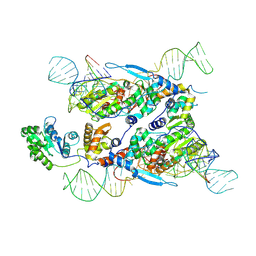 | | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | | Descriptor: | DNA (105-MER), MAGNESIUM ION, RNA (14-MER), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2022-04-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
7V9X
 
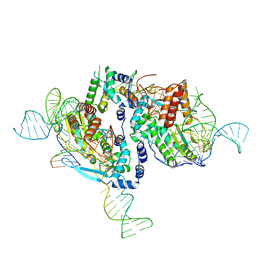 | |
8GQE
 
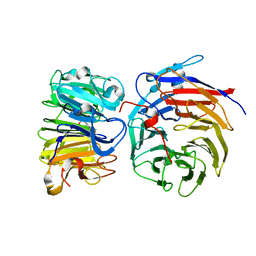 | | Crystal structure of the W285A mutant of UVR8 in complex with RUP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ultraviolet-B receptor UVR8, WD repeat-containing protein RUP2 | | Authors: | Wang, Y.D, Wang, L.X, Guan, Z.Y, chang, H.F, Yin, P. | | Deposit date: | 2022-08-30 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RUP2 facilitates UVR8 redimerization via two interfaces.
Plant Commun., 4, 2023
|
|
5YEJ
 
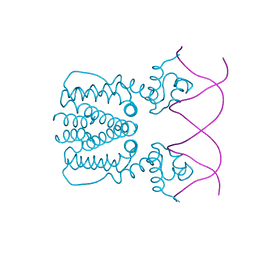 | | Crystal structure of BioQ with its naturel double-stranded DNA operator | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*GP*AP*AP*CP*AP*CP*CP*GP*TP*TP*CP*AP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*TP*GP*AP*AP*CP*GP*GP*TP*GP*TP*TP*CP*AP*GP*GP*T)-3'), TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5YEK
 
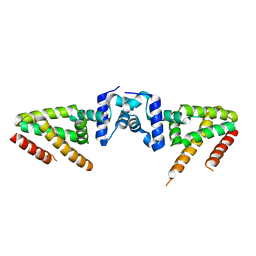 | | Crystal structure of BioQ | | Descriptor: | TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5YGU
 
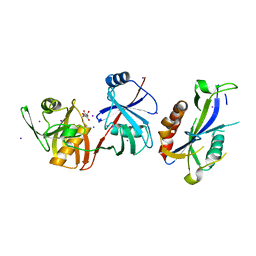 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | Descriptor: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | Deposit date: | 2017-09-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
5ZUG
 
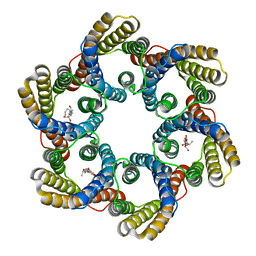 | | Structure of the bacterial acetate channel SatP | | Descriptor: | Succinate-acetate/proton symporter SatP, nonyl beta-D-glucopyranoside | | Authors: | Sun, P.C, Li, J.L, Xiao, Q.J, Guan, Z.Y, Deng, D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of the bacterial acetate transporter SatP reveals that it forms a hexameric channel.
J. Biol. Chem., 293, 2018
|
|
5XYB
 
 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | Descriptor: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | Deposit date: | 2017-07-24 | | Release date: | 2018-09-19 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
8W5J
 
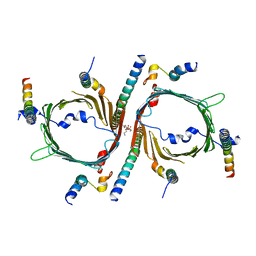 | | Cryo-EM structure of the yeast TOM core complex (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8W5K
 
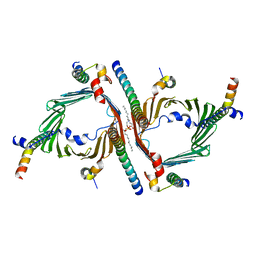 | | Cryo-EM structure of the yeast TOM core complex crosslinked by BS3 (from TOM-TIM23 complex) | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhuang, J.J, Huang, R, Yin, P. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The architecture of substrate-engaged TOM-TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation.
Cell Discov, 10, 2024
|
|
8H0N
 
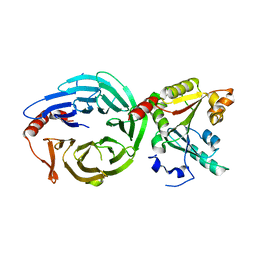 | | Crystal structure of the human METTL1-WDR4 complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Jin, X.H, Guan, Z.Y, Gong, Z, Zhang, D.L. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into how WDR4 promotes the tRNA N7-methylguanosine methyltransferase activity of METTL1.
Cell Discov, 9, 2023
|
|
8HAW
 
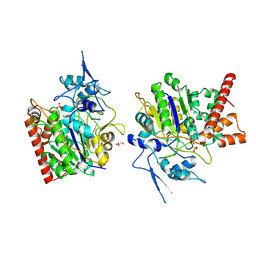 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8HAV
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-specific phospholipase C4 | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
