4ZXC
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Fe3+ | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
4WXO
 
 | |
4WXW
 
 | |
3O72
 
 | | Crystal structure of EfeB in complex with heme | | Descriptor: | OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, Redox component of a tripartite ferrous iron transporter | | Authors: | Liu, X, Du, Q, Wang, Z, Zhu, D, Huang, Y, Li, N, Xu, S, Gu, L. | | Deposit date: | 2010-07-30 | | Release date: | 2011-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes
J.Biol.Chem., 286, 2011
|
|
4ZXD
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD | | Descriptor: | Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
5HHA
 
 | | Structure of PvdO from Pseudomonas aeruginosa | | Descriptor: | CALCIUM ION, PvdO | | Authors: | Bai, G, Yuan, Z, Shang, G, Xia, H, Gu, L. | | Deposit date: | 2016-01-10 | | Release date: | 2017-01-18 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structure of Chromophore maturation protein from Pseudomonas aeruginosa
To Be Published
|
|
4GO4
 
 | | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative gamma-hydroxymuconic semialdehyde dehydrogenase | | Authors: | Su, J, Zhang, C, Liu, S, Zhu, D, Gu, L. | | Deposit date: | 2012-08-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide
To be Published
|
|
4GO3
 
 | |
5ZO4
 
 | | inactive state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
3TLQ
 
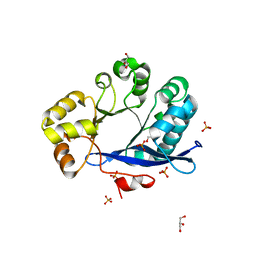 | | Crystal structure of EAL-like domain protein YdiV | | Descriptor: | GLYCEROL, PHOSPHATE ION, Regulatory protein YdiV | | Authors: | Li, B, Li, N, Wang, F, Guo, L, Liu, C, Zhu, D, Xu, S, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
5ZO3
 
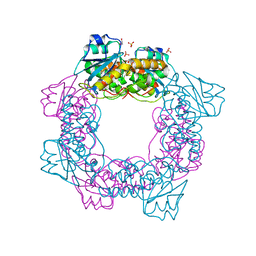 | | apo form of the nuclease | | Descriptor: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
3PFT
 
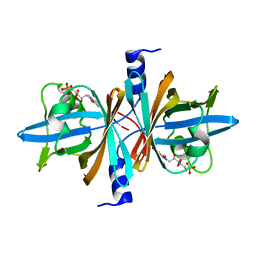 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
3Q7J
 
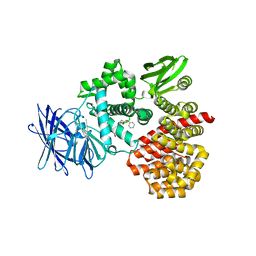 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
2NRF
 
 | | Crystal Structure of GlpG, a Rhomboid family intramembrane protease | | Descriptor: | Protein GlpG | | Authors: | Wu, Z, Yan, N, Feng, L, Yan, H, Gu, L, Shi, Y. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4ZMU
 
 | | Dcsbis, a diguanylate cyclase from Pseudomonas aeruginosa | | Descriptor: | diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | crystal structure of Dcsbis from Pseudomonas aeruginosa
To Be Published
|
|
4ZMM
 
 | | GGDEF domain of Dcsbis complexed with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of Dcsbis GGDEF domain complexed with c-di-GMP
To Be Published
|
|
5XLX
 
 | |
5CET
 
 | | Crystal structure of Rv2837c | | Descriptor: | Bifunctional oligoribonuclease and PAP phosphatase NrnA, MANGANESE (II) ION | | Authors: | Wang, F, He, Q, Zhu, D, Liu, S, Gu, L. | | Deposit date: | 2015-07-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Insight into the Mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
4GFR
 
 | | Crystal Structure of the liganded Chitin Oligasaccharide Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, Peptide ABC transporter, ... | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
3TGV
 
 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
4GF8
 
 | | Crystal Structure of the Chitin Oligasaccharide Binding Protein | | Descriptor: | Peptide ABC transporter, periplasmic peptide-binding protein, SULFATE ION | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
3TLK
 
 | | Crystal structure of holo FepB | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, Ferrienterobactin-binding periplasmic protein, ... | | Authors: | Li, N, Gu, L. | | Deposit date: | 2011-08-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Crystal structure of holo FepB
To be Published
|
|
5XBT
 
 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
5DV9
 
 | |
5DWV
 
 | | Crystal structure of the Luciferase complexed with substrate analogue | | Descriptor: | 2-[6-(cyclobuta-1,3-dien-1-ylamino)-1,3-benzothiazol-2-yl]-1,3-thiazol-4-ol, Luciferin 4-monooxygenase | | Authors: | Su, J, Li, Z, Yuan, Z, Gu, L. | | Deposit date: | 2015-09-23 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Luciferase complexed with substrate analogue
To Be Published
|
|
