4UU3
 
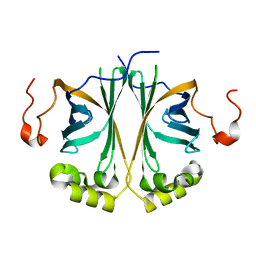 | | Ferulic acid decarboxylase from Enterobacter sp. | | Descriptor: | FERULIC ACID DECARBOXYLASE | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
4V2U
 
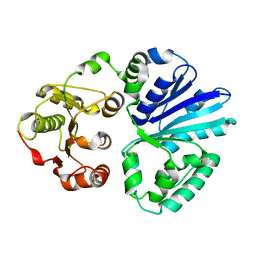 | | Apo-structure of alpha2,3-sialyltransferase from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
7LQU
 
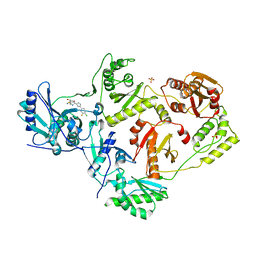 | | Crystal Structure of HIV-1 RT in Complex with NBD-14075 | | Descriptor: | Reverse transcriptase p51, Reverse transcriptase p66, SULFATE ION, ... | | Authors: | Losada, N, Ruiz, F.X, Gruber, K, Das, K, Arnold, E. | | Deposit date: | 2021-02-15 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HIV-1 gp120 Antagonists Also Inhibit HIV-1 Reverse Transcriptase by Bridging the NNRTI and NRTI Sites.
J.Med.Chem., 64, 2021
|
|
2AN7
 
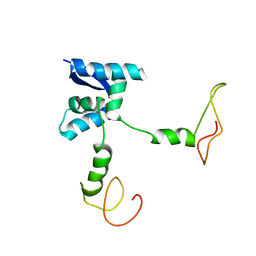 | | Solution structure of the bacterial antidote ParD | | Descriptor: | Protein parD | | Authors: | Oberer, M, Zangger, K, Gruber, K, Keller, W. | | Deposit date: | 2005-08-11 | | Release date: | 2006-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ParD, the antidote of the ParDE toxin antitoxin module, provides the structural basis for DNA and toxin binding.
Protein Sci., 16, 2007
|
|
8PUN
 
 | |
7OUP
 
 | | Structure of human DPP3 in complex with a hydroxyethylene transition state peptidomimetic | | Descriptor: | ((2R,4S,5S)-5-((S)-2-amino-3-methylbutanamido)-2-benzyl-4-hydroxy-6-methylheptanoyl)-L-prolyl-L-tryptophan, Dipeptidyl peptidase 3, MAGNESIUM ION, ... | | Authors: | Kumar, P, Reithofer, V, Gruber, K. | | Deposit date: | 2021-06-12 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Efficient Entropy-Driven Inhibition of Dipeptidyl Peptidase III by Hydroxyethylene Transition-State Peptidomimetics.
Chemistry, 27, 2021
|
|
5M58
 
 | |
7A0Q
 
 | |
7A0T
 
 | |
4BIF
 
 | | Biochemical and structural characterisation of a novel manganese- dependent hydroxynitrile lyase from bacteria | | Descriptor: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | Authors: | Hajnal, I, Lyskowski, A, Hanefeld, U, Gruber, K, Schwab, H, Steiner, K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel Bacterial Manganese-Dependent Hydroxynitrile Lyase.
FEBS J., 280, 2013
|
|
8A85
 
 | |
8P7E
 
 | |
8C66
 
 | |
1JU2
 
 | | Crystal structure of the hydroxynitrile lyase from almond | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dreveny, I, Gruber, K, Glieder, A, Thompson, A, Kratky, C. | | Deposit date: | 2001-08-23 | | Release date: | 2002-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase.
Structure, 9, 2001
|
|
8ADX
 
 | |
8B30
 
 | |
8OIM
 
 | |
8P8F
 
 | |
4V15
 
 | | Crystal structure of D-threonine aldolase from Alcaligenes xylosoxidans | | Descriptor: | D-THREONINE ALDOLASE, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Uhl, M.K, Oberdorfer, G, Steinkellner, G, Riegler, L, Schuermann, M, Gruber, K. | | Deposit date: | 2014-09-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of D-Threonine Aldolase from Alcaligenes Xylosoxidans Provides Insight Into a Metal Ion Assisted Plp-Dependent Mechanism.
Plos One, 10, 2015
|
|
4UU2
 
 | | Ferulic acid decarboxylase from Enterobacter sp., single mutant | | Descriptor: | FERULIC ACID DECARBOXYLASE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
6EZD
 
 | |
4WV8
 
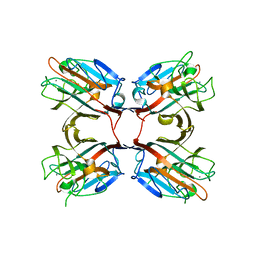 | | Crystal structure of a recombinant Vatairea macrocarpa seed lectin complexed with lactose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Seed lectin, ... | | Authors: | Sousa, B.L, Silva-Filho, J.C, Kumar, P, Lyskowski, A, Bezerra, G.A, Delatorre, P, Rocha, B.A.M, Cunha, R.M.S, Nagano, C.S, Gruber, K, Cavada, B.S. | | Deposit date: | 2014-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a Vatairea macrocarpa lectin in complex with a tumor-associated antigen: A new tool for cancer research.
Int.J.Biochem.Cell Biol., 72, 2016
|
|
5E4D
 
 | |
4ZWN
 
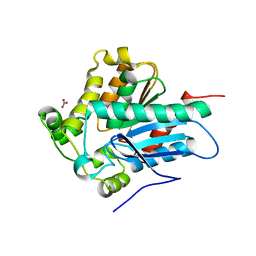 | | Crystal Structure of a Soluble Variant of the Monoglyceride Lipase from Saccharomyces Cerevisiae | | Descriptor: | Monoglyceride lipase, NITRATE ION, SODIUM ION, ... | | Authors: | Aschauer, P, Rengachari, S, Gruber, K, Oberer, M. | | Deposit date: | 2015-05-19 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p.
Biochim.Biophys.Acta, 1861, 2016
|
|
5E4M
 
 | |
