5G4J
 
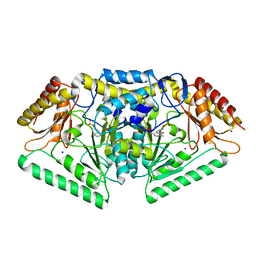 | | Phospholyase A1RDF1 from Arthrobacter in complex with phosphoethanolamine | | Descriptor: | PUTATIVE AMINOTRANSFERASE CLASS III PROTEIN, SODIUM ION, {5-hydroxy-6-methyl-4-[(E)-{[2-(phosphonooxy)ethyl]imino}methyl]pyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
4BMV
 
 | | Short-chain dehydrogenase from Sphingobium yanoikuyae in complex with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SHORT-CHAIN DEHYDROGENASE | | Authors: | Man, H, Kedziora, K, Lavandera-Garcia, I, Gotor-Fernandez, V, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
4ATQ
 
 | | GABA-transaminase A1R958 in complex with external aldimine PLP-GABA adduct | | Descriptor: | 4-AMINOBUTYRATE TRANSAMINASE, GAMMA-AMINO-BUTANOIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Bruce, H, Tuan, A.N, Mangas Sanchez, J, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-05-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of a Gamma-Aminobutyrate (Gaba) Transaminase from the S-Triazine-Degrading Organism Arthrobacter Aurescens Tc1 in Complex with Plp and with its External Aldimine Plp- Gaba Adduct.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4ATP
 
 | | Structure of GABA-transaminase A1R958 from Arthrobacter aurescens in complex with PLP | | Descriptor: | 4-AMINOBUTYRATE TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Bruce, H, Tuan, A.N, Mangas Sanchez, J, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-05-09 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a Gamma-Aminobutyrate (Gaba) Transaminase from the S-Triazine-Degrading Organism Arthrobacter Aurescens Tc1 in Complex with Plp and with its External Aldimine Plp- Gaba Adduct.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4D3S
 
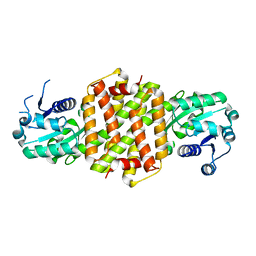 | | Imine reductase from Nocardiopsis halophila | | Descriptor: | IMINE REDUCTASE, octyl beta-D-glucopyranoside | | Authors: | Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-10-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, Activity and Stereoselectivity of Nadph-Dependent Oxidoreductases Catalysing the S-Selective Reduction of the Imine Substrate 2-Methylpyrroline.
Chembiochem, 16, 2015
|
|
6S5J
 
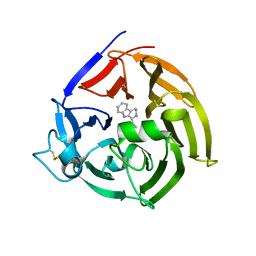 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-Ethyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-ethyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5Q
 
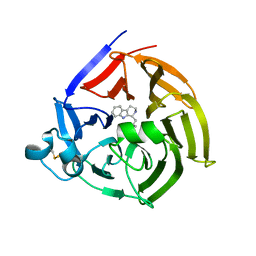 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-isobutyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-(2-methylpropyl)-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5U
 
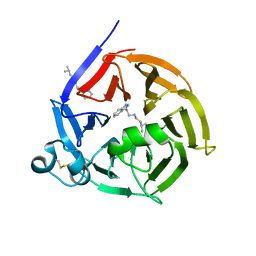 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with N-[2-(1H-Indol-3-yl)ethyl]-3-methyl-1-butanamine | | Descriptor: | Strictosidine synthase, ~{N}-[2-(1~{H}-indol-3-yl)ethyl]-3-methyl-butan-1-amine | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6S5M
 
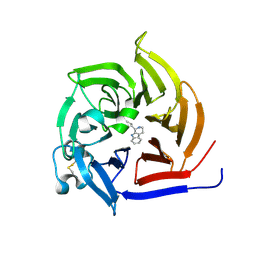 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with (S)-1-n-propyl-2,3,4,9-tetrahydro-1H-beta-carboline | | Descriptor: | (1~{S})-1-propyl-2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Strictosidine synthase | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
6SQ8
 
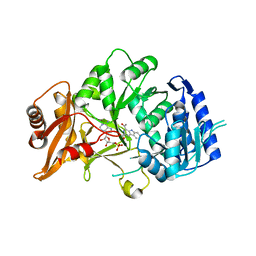 | | Structure of amide bond synthetase McbA from Marinactinospora thermotolerans | | Descriptor: | 1-ethanoyl-9~{H}-pyrido[3,4-b]indole-3-carboxylic acid, ADENOSINE MONOPHOSPHATE, Fatty acid CoA ligase | | Authors: | Rowlinson, B, Petchey, M, Grogan, G. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biocatalytic Synthesis of Moclobemide Using the Amide Bond Synthetase McbA Coupled with an ATP Recycling System.
Acs Catalysis, 10, 2020
|
|
6TFN
 
 | |
6SKX
 
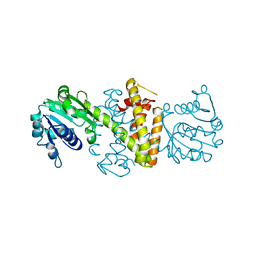 | | Structure of Reductive Aminase from Neosartorya fumigata | | Descriptor: | Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6THM
 
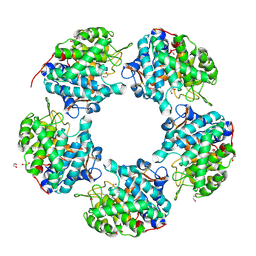 | | Linalool Dehydratase Isomerase M125A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI, MALONATE ION | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
6T9H
 
 | |
6SLE
 
 | | Structure of Reductive Aminase from Neosartorya fumigata in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6TFT
 
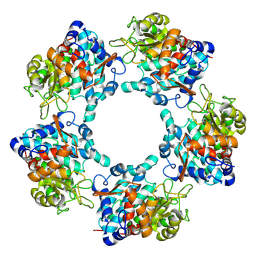 | |
6TFR
 
 | | Linalool Dehydratase Isomerase C180A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
6TOE
 
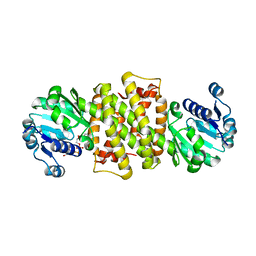 | |
6TO4
 
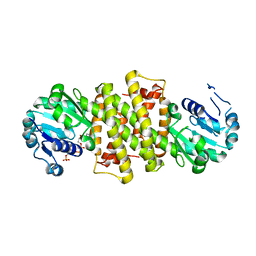 | |
