3ZGJ
 
 | |
4A6G
 
 | | N-acyl amino acid racemase from Amycalotopsis sp. Ts-1-60: G291D- F323Y mutant in complex with N-acetyl methionine | | Descriptor: | MAGNESIUM ION, N-ACETYLMETHIONINE, N-ACYLAMINO ACID RACEMASE | | Authors: | Baxter, S, Royer, S, Grogan, G, Holt-Tiffin, K.E, Taylor, I.N, Fotheringham, I.G, Campopiano, D.J. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | An Improved Racemase/Acylase Biotransformation for the Preparation of Enantiomerically Pure Amino Acids.
J.Am.Chem.Soc., 134, 2012
|
|
2WQ8
 
 | | Glycan labelling using engineered variants of galactose oxidase obtained by directed evolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Rannes, J.G, Ioannou, A, Willies, S.C, Behrens, C, Grogan, G.J, Flitsch, S.L, Turner, N.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Glycoprotein Labeling Using Engineered Variants of Galactose Oxidase Obtained by Directed Evolution.
J.Am.Chem.Soc., 133, 2011
|
|
5A5K
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with camalexin | | Descriptor: | (2Z)-2-indol-3-ylidene-3H-1,3-thiazole, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-18 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A9R
 
 | | Apo form of Imine reductase from Amycolatopsis orientalis | | Descriptor: | ACETATE ION, IMINE REDUCTASE | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2015-07-22 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
6IAU
 
 | | Amine Dehydrogenase from Cystobacter fuscus in complex with NADP+ and cyclohexylamine | | Descriptor: | Amine Dehydrogenase, CYCLOHEXYLAMMONIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Beloti, L, Mayol, O, Turkenburg, J.P, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
5A9T
 
 | | Imine Reductase from Amycolatopsis orientalis in complex with (R)- Methyltetrahydroisoquinoline | | Descriptor: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, ACETATE ION, CALCIUM ION, ... | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2015-07-22 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
6EOD
 
 | |
6EOH
 
 | | Reductive Aminase from Aspergillus terreus in complex with NADPH and ethyl levulinate | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductive Aminase, ethyl levulinate | | Authors: | Sharma, M, Grogan, G, Mangas-Sanchez, J, Turner, N.J. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mechanism for Reductive Amination Catalyzed by Fungal Reductive Aminases
Acs Catalysis, 2018
|
|
6EOI
 
 | | Reductive Aminase from Aspergillus terreus in complex with NADPH and ethyl-5-oxohexanoate | | Descriptor: | MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductive Aminase, ... | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Mechanism for Reductive Amination Catalyzed by Fungal Reductive Aminases
Acs Catalysis, 2018
|
|
5A4U
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with indole-3-aldehyde | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
6G1M
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open and closed form | | Descriptor: | Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Beloti, L, Frese, A, Mayol, O, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
5A4V
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
5A4W
 
 | | AtGSTF2 from Arabidopsis thaliana in complex with quercetrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, ACETATE ION, GLUTATHIONE S-TRANSFERASE F2 | | Authors: | Ahmad, L, Rylott, E, Bruce, N.C, Edwards, R, Grogan, G. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands.
FEBS Open Bio, 7, 2017
|
|
6GIO
 
 | |
2VVL
 
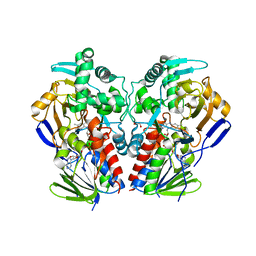 | | The structure of MAO-N-D3, a variant of monoamine oxidase from Aspergillus niger. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
3ZHB
 
 | |
3ZGY
 
 | |
6G71
 
 | | Structure of CYP1232A24 from Arthrobacter sp. | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, FE (III) ION, ... | | Authors: | Dubiel, P, Sharma, M, Klenk, J, Hauer, B, Grogan, G. | | Deposit date: | 2018-04-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of cytochrome P450 1232A24 and 1232F1 from Arthrobacter sp. and their role in the metabolic pathway of papaverine.
J.Biochem., 166, 2019
|
|
3ZDN
 
 | | D11-C mutant of monoamine oxidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Frank, A, Ghislieri, D, Willies, S, Turner, N.J, Grogan, G. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineering an Enantioselective Amine Oxidase for the Synthesis of Pharmaceutical Building Blocks and Alkaloid Natural Products.
J.Am.Chem.Soc., 135, 2013
|
|
4ALB
 
 | | Structure of Phenolic Acid Decarboxylase from Bacillus subtilis: Tyr19Ala mutant in complex with coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHENOLIC ACID DECARBOXYLASE PADC | | Authors: | Frank, A, Eborall, W, Hyde, R, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-03-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mutational Analysis of Phenolic Acid Decarboxylase from Bacillus Subtilis (Bspad), which Converts Bio-Derived Phenolic Acids to Styrene Derivatives
Catal.Sci.Technol., 2, 2012
|
|
2XAA
 
 | | Alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541 at pH 8.5 in complex with NAD and butane-1,4-diol | | Descriptor: | 1,4-BUTANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SECONDARY ALCOHOL DEHYDROGENASE, ... | | Authors: | Kroutil, W, Gruber, K, Grogan, G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Substrate Specificity and Solvent Tolerance in Alcohol Dehydrogenase Adh-'A' from Rhodococcus Ruber Dsm 44541.
Chem.Commun.(Camb.), 46, 2010
|
|
4A9W
 
 | | Flavin-containing monooxygenase from Stenotrophomonas maltophilia | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MONOOXYGENASE, SULFATE ION | | Authors: | Jensen, C.N, Cartwright, J, Hart, S, Turkenburg, J.P, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2011-11-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Flavoprotein Monooxygenase that Catalyses a Baeyer-Villiger Reaction and Thioether Oxidation Using Nadh as the Nicotinamide Cofactor.
Chembiochem, 13, 2012
|
|
2WIY
 
 | | Cytochrome P450 XplA heme domain P21212 | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450-LIKE PROTEIN XPLA, IMIDAZOLE, ... | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
