6PF0
 
 | |
6PEZ
 
 | |
5Z6O
 
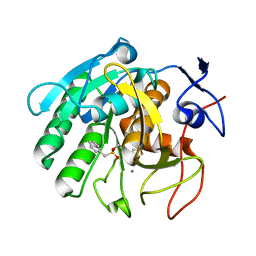 | | Crystal structure of Penicillium cyclopium protease | | Descriptor: | CALCIUM ION, phenylmethanesulfonic acid, protease | | Authors: | Ko, T.-P, Koszelak, S, Ng, J, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystallographic structure of the subtilisin protease from Penicillium cyclopium.
Biochemistry, 36, 1997
|
|
1IAI
 
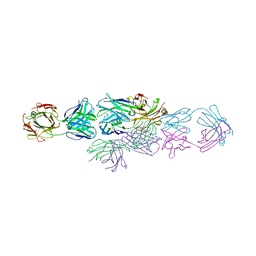 | | IDIOTYPE-ANTI-IDIOTYPE FAB COMPLEX | | Descriptor: | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A), IDIOTYPIC FAB 730.1.4 (IGG1) OF VIRUS NEUTRALIZING ANTIBODY | | Authors: | Ban, N, Escobar, C, Garcia, R, Hasel, K, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-12-28 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of an idiotype-anti-idiotype Fab complex.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1AIF
 
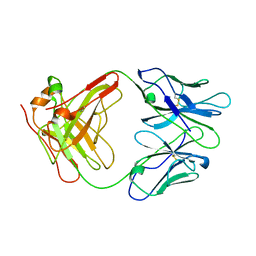 | | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB FROM MOUSE | | Descriptor: | ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (HEAVY CHAIN), ANTI-IDIOTYPIC FAB 409.5.3 (IGG2A) FAB (LIGHT CHAIN) | | Authors: | Ban, N, Escobar, C, Hasel, K, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-11-14 | | Release date: | 1997-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an anti-idiotypic Fab against feline peritonitis virus-neutralizing antibody and a comparison with the complexed Fab.
FASEB J., 9, 1995
|
|
1A34
 
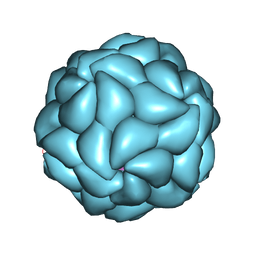 | | SATELLITE TOBACCO MOSAIC VIRUS/RNA COMPLEX | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), SATELLITE TOBACCO MOSAIC VIRUS, ... | | Authors: | Larson, S.B, Day, J, Greenwood, A.J, McPherson, A. | | Deposit date: | 1998-01-28 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Refined structure of satellite tobacco mosaic virus at 1.8 A resolution.
J.Mol.Biol., 277, 1998
|
|
1DDL
 
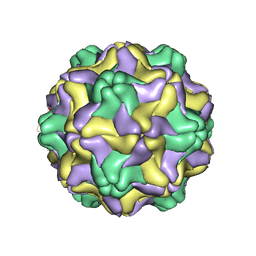 | | DESMODIUM YELLOW MOTTLE TYMOVIRUS | | Descriptor: | DESMODIUM YELLOW MOTTLE VIRUS, RNA (5'-R(P*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Larson, S.B, Day, J, Canady, M.A, Greenwood, A, McPherson, A. | | Deposit date: | 1999-11-10 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Refined structure of desmodium yellow mottle tymovirus at 2.7 A resolution.
J.Mol.Biol., 301, 2000
|
|
1CAW
 
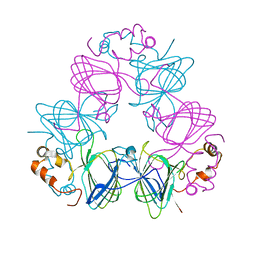 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-06-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1CAU
 
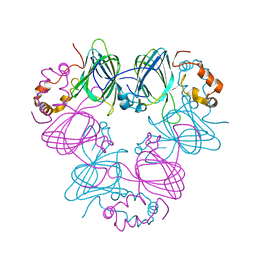 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-07-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1THV
 
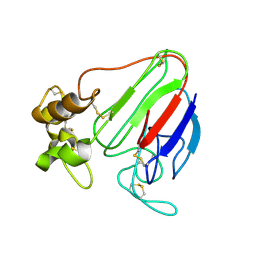 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | THAUMATIN ISOFORM A | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1THW
 
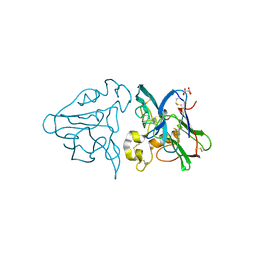 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1CAX
 
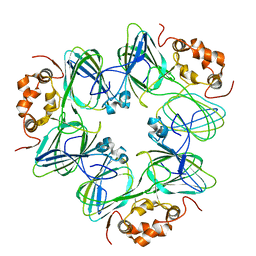 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1THU
 
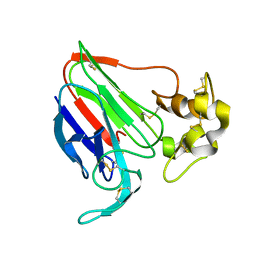 | | THE STRUCTURES OF THREE CRYSTAL FORMS OF THE SWEET PROTEIN THAUMATIN | | Descriptor: | THAUMATIN ISOFORM B | | Authors: | Ko, T.-P, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1994-06-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of three crystal forms of the sweet protein thaumatin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
