4PGG
 
 | | Caffeic acid O-methyltransferase from Sorghum bicolor | | Descriptor: | Caffeic acid O-methyltransferase | | Authors: | Green, A.R, Lewis, K.M, Kang, C. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations.
Plant Physiol., 165, 2014
|
|
4PGH
 
 | | Caffeic acid O-methyltransferase from Sorghum bicolor | | Descriptor: | Caffeic acid O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Green, A.R, Lewis, K.M, Kang, C. | | Deposit date: | 2014-05-02 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Determination of the Structure and Catalytic Mechanism of Sorghum bicolor Caffeic Acid O-Methyltransferase and the Structural Impact of Three brown midrib12 Mutations.
Plant Physiol., 165, 2014
|
|
4FQU
 
 | |
4G0L
 
 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G0I
 
 | | Glutathionyl-Hydroquinone Reductase, YqjG of Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, protein yqjG | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G0K
 
 | | Glutathionyl-hydroquinone reductase, YqjG, of E.coli complexed with GS-menadione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-gamma-glutamyl-S-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-L-cysteinylglycine, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
6PF0
 
 | |
6PEZ
 
 | |
4XIV
 
 | |
6H08
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a His175Me-His proximal ligand substitution | | Descriptor: | COBALT (II) ION, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2018-07-06 | | Release date: | 2020-02-12 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rewiring the "Push-Pull" Catalytic Machinery of a Heme Enzyme Using an Expanded Genetic Code.
Acs Catalysis, 10, 2020
|
|
7QVH
 
 | |
4LZI
 
 | | Characterization of Solanum tuberosum Multicystatin and Significance of Core Domains | | Descriptor: | Multicystatin | | Authors: | Nissen, M.S, Kumar, G.N, Green, A.R, Knowles, N.R, Kang, C. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Solanum tuberosum Multicystatin and the Significance of Core Domains.
Plant Cell, 25, 2013
|
|
6Y2Y
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with Trp51 to S-Trp51 and Trp191Phe modifications | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
6Y1T
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
5OJA
 
 | | Structure of MbQ | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJB
 
 | | Structure of MbQ NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJC
 
 | | Structure of MbQ2.1 NMH | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
5OJ9
 
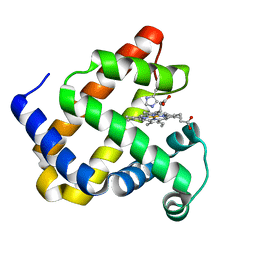 | | Structure of Mb NMH | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, T, Pott, M, Mori, T, Mittl, P, Green, A, Hivert, D. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | A Noncanonical Proximal Heme Ligand Affords an Efficient Peroxidase in a Globin Fold.
J. Am. Chem. Soc., 140, 2018
|
|
6BAS
 
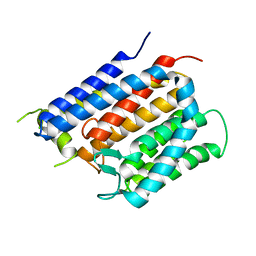 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
4HUZ
 
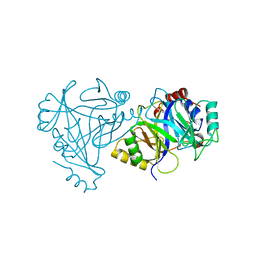 | | 2,6-Dichloro-p-hydroquinone 1,2-Dioxygenase | | Descriptor: | 2,6-dichloro-p-hydroquinone 1,2-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Hayes, R.P, Nissen, M.S, Green, A.R, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) from Sphingobium chlorophenolicum, a new type of aromatic ring-cleavage enzyme.
Mol.Microbiol., 88, 2013
|
|
1SZE
 
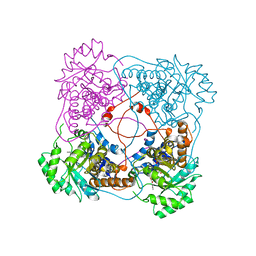 | | L230A mutant flavocytochrome b2 with benzoylformate | | Descriptor: | BENZOYL-FORMIC ACID, Cytochrome b2, mitochondrial, ... | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
1SZG
 
 | | A198G:L230A flavocytochrome b2 with sulfite bound | | Descriptor: | Cytochrome b2, mitochondrial, N-SULFO-FLAVIN MONONUCLEOTIDE | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
1SZF
 
 | | A198G:L230A mutant flavocytochrome b2 with pyruvate bound | | Descriptor: | Cytochrome b2, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mowat, C.G, Wehenkel, A, Green, A.J, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2004-04-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Altered Substrate Specificity in Flavocytochrome b(2): Structural Insights into the Mechanism of l-Lactate Dehydrogenation
Biochemistry, 43, 2004
|
|
5Z6O
 
 | | Crystal structure of Penicillium cyclopium protease | | Descriptor: | CALCIUM ION, phenylmethanesulfonic acid, protease | | Authors: | Ko, T.-P, Koszelak, S, Ng, J, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystallographic structure of the subtilisin protease from Penicillium cyclopium.
Biochemistry, 36, 1997
|
|
