1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
4NYK
 
 | | Structure of a membrane protein | | Descriptor: | Acid-sensing ion channel 1, CHLORIDE ION | | Authors: | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pore architecture and ion sites in acid-sensing ion channels and P2X receptors.
Nature, 460, 2009
|
|
1MM7
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Quisqualate in a Zinc Crystal Form at 1.65 Angstroms Resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Horning, M, Mayer, M.L, Gouaux, E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-02-04 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of Activation and Selectivity in a Ligand-Gated Ion Channel: Structural and Functional Studies of GluR2 and Quisqualate
Biochemistry, 41, 2003
|
|
1MM6
 
 | | crystal structure of the GluR2 ligand binding core (S1S2J) in complex with quisqualate in a non zinc crystal form at 2.15 angstroms resolution | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, GLYCEROL, ... | | Authors: | Jin, R, Horning, M, Mayer, M.L, Gouaux, E. | | Deposit date: | 2002-09-03 | | Release date: | 2003-02-04 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of activation and selectivity in a ligand-gated ion channel: Structural and functional studies of GluR2 and quisqualate
Biochemistry, 41, 2002
|
|
3USG
 
 | | Crystal structure of LeuT bound to L-leucine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, LEUCINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3USJ
 
 | |
3USK
 
 | |
3USL
 
 | | Crystal Structure of LeuT bound to L-selenomethionine in space group C2 from lipid bicelles | | Descriptor: | ACETATE ION, IODIDE ION, PHOSPHOCHOLINE, ... | | Authors: | Wang, H, Elferich, J, Gouaux, E. | | Deposit date: | 2011-11-23 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8DE4
 
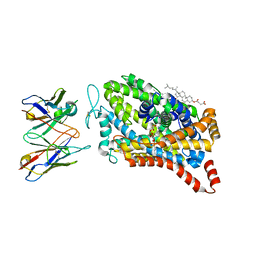 | |
8DE3
 
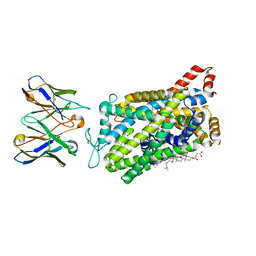 | |
3H5W
 
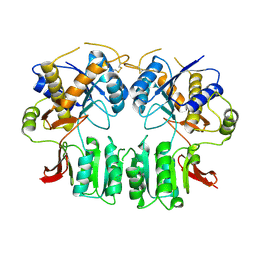 | | Crystal structure of the GluR2-ATD in space group P212121 without solvent | | Descriptor: | Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
3H9V
 
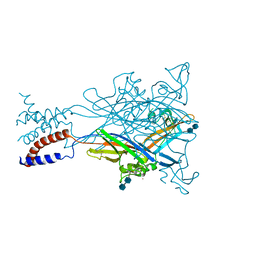 | | Crystal structure of the ATP-gated P2X4 ion channel in the closed, apo state at 3.1 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GADOLINIUM ATOM, P2X purinoceptor | | Authors: | Kawate, T, Michel, J.C, Gouaux, E. | | Deposit date: | 2009-04-30 | | Release date: | 2009-07-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the ATP-gated P2X(4) ion channel in the closed state.
Nature, 460, 2009
|
|
3H5V
 
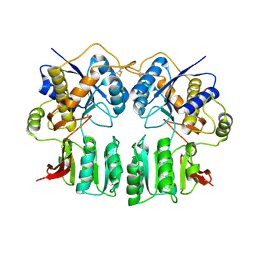 | | Crystal structure of the GluR2-ATD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Jin, R, Singh, S.K, Gu, S, Furukawa, H, Sobolevsky, A, Zhou, J, Jin, Y, Gouaux, E. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and association behaviour of the GluR2 amino-terminal domain.
Embo J., 28, 2009
|
|
1MQJ
 
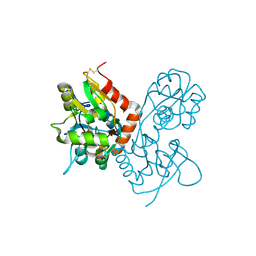 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MQG
 
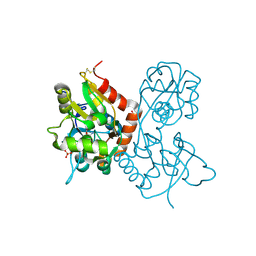 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1MXX
 
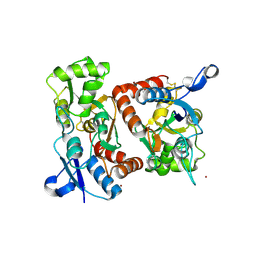 | | crystal titration experiments (AMPA co-crystals soaked in 100 uM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MY3
 
 | | crystal structure of glutamate receptor ligand-binding core in complex with bromo-willardiine in the Zn crystal form | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core:
Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
1MY0
 
 | | crystal titration experiments (AMPA co-crystals soaked in 100 nM BrW) | | Descriptor: | GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Function, Conformational Plasticity, and Dimer-Dimer Contacts of the GluR2 Ligand-Binding Core: Studies of 5-Substituted Willardiines and GluR2 S1S2 in the Crystal
Biochemistry, 42, 2003
|
|
2AL4
 
 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | Authors: | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
7USY
 
 | | Structure of C. elegans TMC-1 complex with ARRD-6 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USW
 
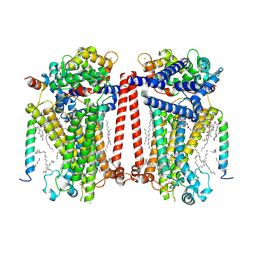 | | Structure of Expanded C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USX
 
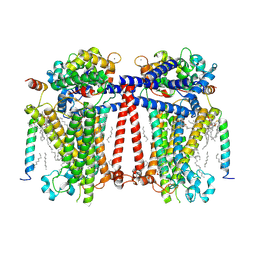 | | Structure of Contracted C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7LGU
 
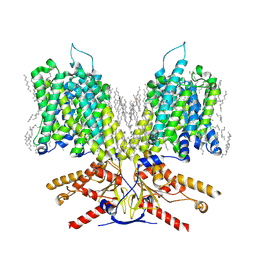 | |
7LGW
 
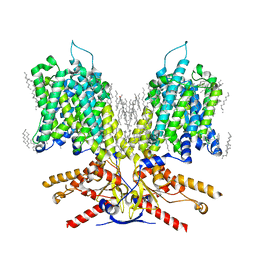 | |
