4NNC
 
 | | Ternary complex of ObcA with C4-CoA adduct and oxalate | | Descriptor: | (3S)-3-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-3-oxidanyl-butanoic acid, COBALT (II) ION, OBCA, ... | | Authors: | Oh, J.T, Goo, E, Hwang, I, Rhee, S. | | Deposit date: | 2013-11-17 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structural Basis for Bacterial Quorum Sensing-mediated Oxalogenesis.
J.Biol.Chem., 289, 2014
|
|
4NNA
 
 | | Apo structure of ObcA | | Descriptor: | MAGNESIUM ION, OBCA, Oxalate Biosynthetic Component A | | Authors: | Oh, J.T, Goo, E, Hwang, I, Rhee, S. | | Deposit date: | 2013-11-17 | | Release date: | 2014-03-19 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural Basis for Bacterial Quorum Sensing-mediated Oxalogenesis.
J.Biol.Chem., 289, 2014
|
|
4NNB
 
 | | Binary complex of ObcA with oxaloacetate | | Descriptor: | MAGNESIUM ION, OBCA, Oxalate Biosynthetic Component A, ... | | Authors: | Oh, J.T, Goo, E, Hwang, I, Rhee, S. | | Deposit date: | 2013-11-17 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Bacterial Quorum Sensing-mediated Oxalogenesis.
J.Biol.Chem., 289, 2014
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
8QYS
 
 | | Human preholo proteasome 20S core particle | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYJ
 
 | | Human 20S proteasome assembly structure 1 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome assembly chaperone 3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYO
 
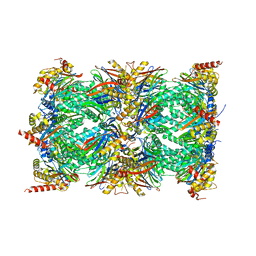 | | Human proteasome 20S core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QZ9
 
 | | Human 20S proteasome assembly intermediate structure 4 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYL
 
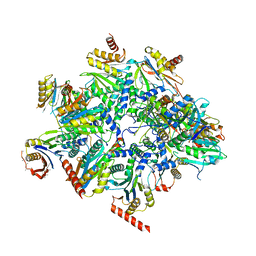 | | Human 20S proteasome assembly intermediate structure 2 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYM
 
 | | Human 20S proteasome assembly intermediate structure 3 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYN
 
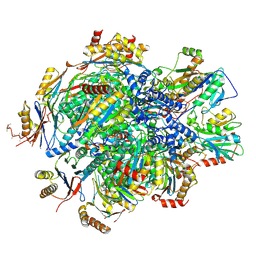 | | Human 20S proteasome assembly intermediate structure 5 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1CM3
 
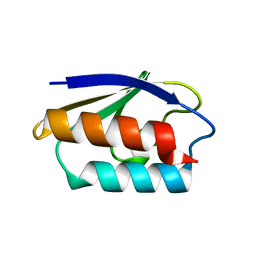 | | HIS15ASP HPR FROM E. COLI | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Waygood, E.B, Delbaere, L.T.J. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
1CM2
 
 | | STRUCTURE OF HIS15ASP HPR AFTER HYDROLYSIS OF RINGED SPECIES. | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Delbaere, L.T.J, Waygood, E.B. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
2JEL
 
 | | JEL42 FAB/HPR COMPLEX | | Descriptor: | HISTIDINE-CONTAINING PROTEIN, JEL42 FAB FRAGMENT, SULFATE ION | | Authors: | Prasad, L, Waygood, E.B, Lee, J.S, Delbaere, L.T.J. | | Deposit date: | 1998-02-24 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A resolution structure of the jel42 Fab fragment/HPr complex
J.Mol.Biol., 280, 1998
|
|
6EF0
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (1D* motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF1
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.73 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF2
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (5T motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
6EF3
 
 | | Yeast 26S proteasome bound to ubiquitinated substrate (4D motor state) | | Descriptor: | 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, 26S proteasome regulatory subunit 6B homolog, ... | | Authors: | de la Pena, A.H, Goodall, E.A, Gates, S.N, Lander, G.C, Martin, A. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Substrate-engaged 26Sproteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation.
Science, 362, 2018
|
|
