5MF2
 
 | | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 | | Descriptor: | CHLORIDE ION, Distal tail protein, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Lassalle, L, Arnaud, C.-A, Breyton, C, Madern, D, Coquelle, N, Maury, O, Girard, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence.
Chem Sci, 8, 2017
|
|
7QO8
 
 | |
2Y39
 
 | | Ni-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | ACETATE ION, NICKEL (II) ION, NICKEL AND COBALT RESISTANCE PROTEIN CNRR | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3H
 
 | | E63Q mutant of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3B
 
 | | Co-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | COBALT (II) ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.554 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3D
 
 | | Zn-bound form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ZINC ION | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
2Y3G
 
 | | Se-Met form of Cupriavidus metallidurans CH34 CnrXs | | Descriptor: | CHLORIDE ION, GLYCEROL, NICKEL AND COBALT RESISTANCE PROTEIN CNRR, ... | | Authors: | Trepreau, J, Girard, E, Maillard, A.P, de Rosny, E, Petit-Haertlein, I, Kahn, R, Coves, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural Basis for Metal Sensing by Cnrx.
J.Mol.Biol., 408, 2011
|
|
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8AB3
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in complex with oxamate, NADH and FBP. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,6-di-O-phosphono-beta-D-fructofuranose, L-lactate dehydrogenase, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8Q8O
 
 | |
6Q3T
 
 | | Structure of Protease1 from Pyrococcus horikoshii at room temperature in ChipX microfluidic device | | Descriptor: | Deglycase PH1704 | | Authors: | de Wijn, R, Engilberge, S, Olieric, V, Girard, E, Sauter, C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6QSS
 
 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
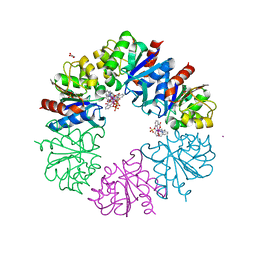
6F2I
 
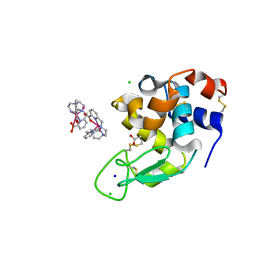 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Engilberge, S, Riobe, F, DI Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6F2K
 
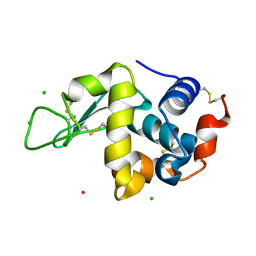 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM potassium phosphate monobasic. | | Descriptor: | CHLORIDE ION, Lysozyme C, TERBIUM(III) ION | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6F2M
 
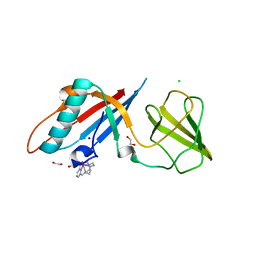 | | Structure of the bacteriophage T5 distal tail protein pb9 co-crystallized with 10mM Tb-Xo4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Distal tail protein, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Breyton, C, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6F2F
 
 | | Crystal structure of Protease 1 from Pyrococcus Horikoshii co-cystallized in presence of 10 mM Tb-Xo4 and ammonium sulfate. | | Descriptor: | Deglycase PH1704, Dihydroxyacetone, SULFATE ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Franzetti, B, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6F2J
 
 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM sodium sulfate | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
4BAF
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethyltriazoledipicolinate complex at 1.51 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
5LDA
 
 | | Structure of deubiquitinating enzyme homolog (Pyrococcus furiosus JAMM1) in complex with ubiquitin-like SAMP2. | | Descriptor: | GLYCEROL, JAMM1, SAMP2, ... | | Authors: | Cao, S, Engilberge, S, Girard, E, Gabel, F, Franzetti, B, Maupin-Furlow, J.A. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
5LD9
 
 | | Structure of deubiquitinating enzyme homolog, Pyrococcus furiosus JAMM1. | | Descriptor: | CHLORIDE ION, JAMM1, ZINC ION | | Authors: | Maupin-Furlow, J.A, Franzetti, B, Cao, S, Girard, E, Gabel, F, Engilberge, S. | | Deposit date: | 2016-06-24 | | Release date: | 2017-05-17 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
4BAD
 
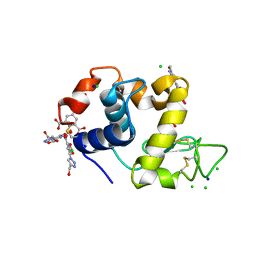 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxymethyltriazoledipicolinate complex at 1.35 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4BAR
 
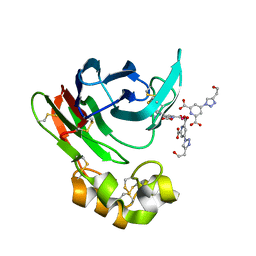 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxyethyltriazoledipicolinate complex at 1.20 A resolution. | | Descriptor: | 4-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Clicked europium dipicolinate complexes for protein X-ray structure determination.
Chem. Commun. (Camb.), 48, 2012
|
|
4BAP
 
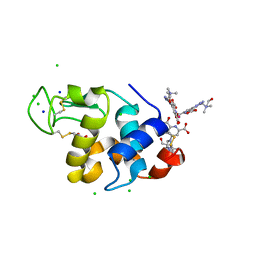 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxyethylcholinetriazoledipicolinate complex at 1.21 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, EUROPIUM (III) ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4BAL
 
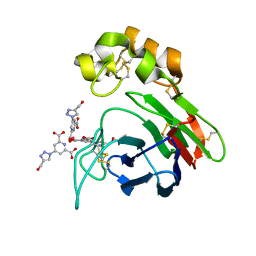 | | Thaumatin from Thaumatococcus daniellii structure in complex with the europium tris-hydroxymethyltriazoledipicolinate complex at 1.30 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, EUROPIUM (III) ION, THAUMATIN-1 | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
5NGJ
 
 | | Crystal structure of pb6, major tail tube protein of bacteriophage T5 | | Descriptor: | CHLORIDE ION, Tail tube protein | | Authors: | Arnaud, C.-A, Effantin, G, Vives, C, Engilberge, S, Bacia, M, Boulanger, P, Girard, E, Schoehn, G, Breyton, C. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacteriophage T5 tail tube structure suggests a trigger mechanism for Siphoviridae DNA ejection.
Nat Commun, 8, 2017
|
|
