3L7E
 
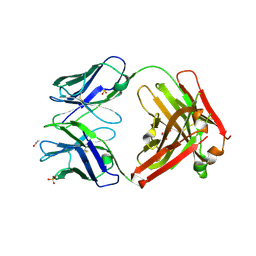 | | Crystal structure of ANTI-IL-13 antibody C836 | | Descriptor: | ACETATE ION, C836 HEAVY CHAIN, C836 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen recognition by antibody C836 through adjustment of VL/VH packing
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1R4B
 
 | |
1S4C
 
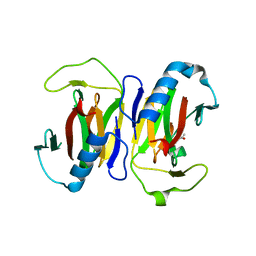 | | YHCH PROTEIN (HI0227) COPPER COMPLEX | | Descriptor: | ACETATE ION, COPPER (II) ION, Protein HI0227 | | Authors: | Teplyakov, A, Obmolova, G, Toedt, J, Gilliland, G.L. | | Deposit date: | 2004-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism.
J.Bacteriol., 187, 2005
|
|
1R45
 
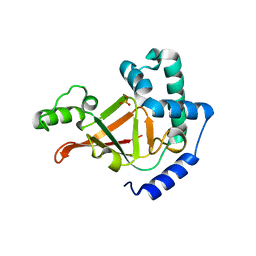 | | ADP-ribosyltransferase C3bot2 from Clostridium botulinum, triclinic form | | Descriptor: | GLYCEROL, Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L, Narumiya, S. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of ADP-ribosyltransferase C3bot2 from Clostridium botulinum
To be Published
|
|
1R4W
 
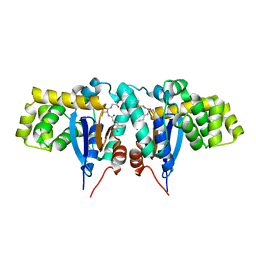 | | Crystal structure of Mitochondrial class kappa glutathione transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, mitochondrial | | Authors: | Ladner, J.E, Parsons, J.F, Rife, C.L, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2003-10-08 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Parallel Evolutionary Pathways for Glutathione Transferases: Structure and Mechanism of the Mitochondrial Class Kappa Enzyme rGSTK1-1
Biochemistry, 43, 2004
|
|
1SUA
 
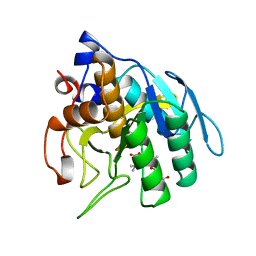 | | SUBTILISIN BPN' | | Descriptor: | SUBTILISIN BPN', TETRAPEPTIDE ALA-LEU-ALA-LEU | | Authors: | Almog, O, Gilliland, G.L. | | Deposit date: | 1997-01-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of calcium-independent subtilisin BPN' with restored thermal stability folded without the prodomain.
Proteins, 31, 1998
|
|
1RW1
 
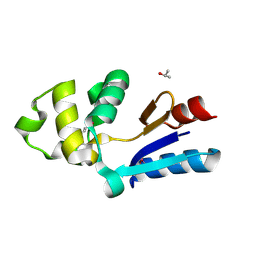 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1SUC
 
 | |
1SUE
 
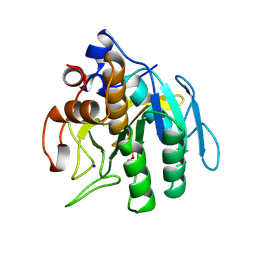 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
1CLS
 
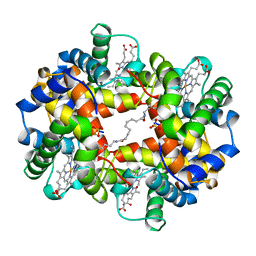 | | CROSS-LINKED HUMAN HEMOGLOBIN DEOXY | | Descriptor: | HEMOGLOBIN, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ji, X, Fronticelli, C, Bucci, E, Gilliland, G.L. | | Deposit date: | 1995-08-29 | | Release date: | 1996-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Positive and negative cooperativities at subsequent steps of oxygenation regulate the allosteric behavior of multistate sebacylhemoglobin.
Biochemistry, 35, 1996
|
|
1SUD
 
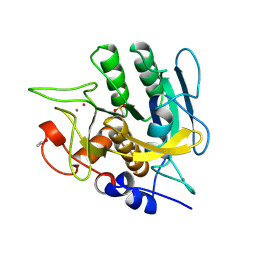 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
1SUP
 
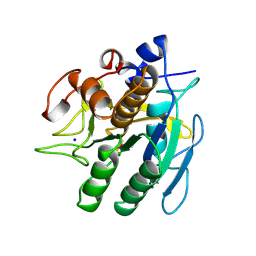 | | SUBTILISIN BPN' AT 1.6 ANGSTROMS RESOLUTION: ANALYSIS OF DISCRETE DISORDER AND COMPARISON OF CRYSTAL FORMS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN BPN', ... | | Authors: | Gallagher, D.T, Oliver, J.D, Betzel, C, Gilliland, G.L. | | Deposit date: | 1995-08-14 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Subtilisin BPN' at 1.6 A resolution: analysis for discrete disorder and comparison of crystal forms.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1SUB
 
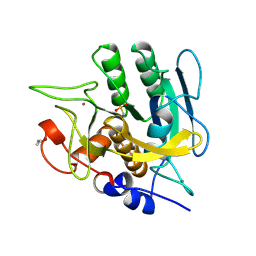 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
4LPT
 
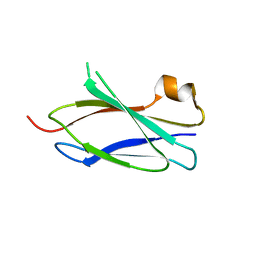 | |
4M7K
 
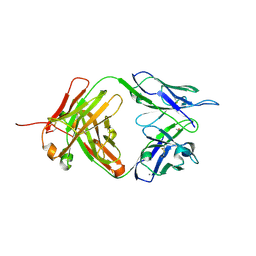 | | Crystal structure of anti-tissue factor antibody 10H10 | | Descriptor: | 10H10 heavy chain, 10H10 light chain, ACETATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4MA3
 
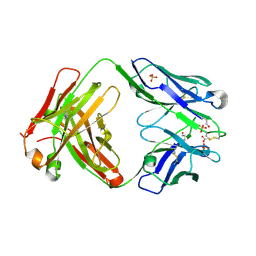 | | Crystal structure of anti-hinge rabbit antibody C2095 | | Descriptor: | ACETATE ION, C2095 heavy chain, C2095 light chain, ... | | Authors: | Malia, T, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-08-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
4LPX
 
 | |
4M6M
 
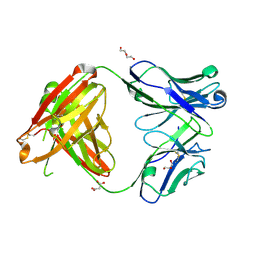 | | Crystal structure of anti-IL-23 antibody CNTO1959 at pH 9.5 | | Descriptor: | CNTO1959 heavy chain, CNTO1959 light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4KUZ
 
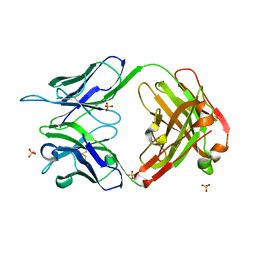 | |
4LPW
 
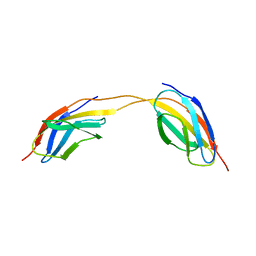 | |
4M6A
 
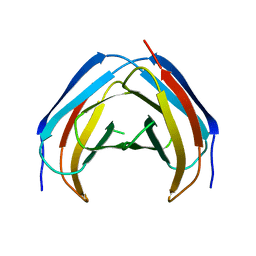 | | N-Terminal beta-Strand Swapping in a Consensus Derived Alternative Scaffold Driven by Stabilizing Hydrophobic Interactions | | Descriptor: | Tencon | | Authors: | Luo, J, Teplyakov, A, Obmolova, G, Malia, T.J, Chan, W, Jocobs, S.A, O'neil, K.T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | N-terminal beta-strand swapping in a consensus-derived alternative scaffold driven by stabilizing hydrophobic interactions.
Proteins, 82, 2014
|
|
4M6O
 
 | | Crystal structure of anti-NGF antibody CNTO7309 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO7309 heavy chain, CNTO7309 light chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4MAU
 
 | | Crystal structure of anti-ST2L antibody C2244 | | Descriptor: | C2244 heavy chain, C2244 light chain, FORMIC ACID, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
1AJ9
 
 | | R-STATE HUMAN CARBONMONOXYHEMOGLOBIN ALPHA-A53S | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), ... | | Authors: | Vasquez, G.B, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1998-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human carboxyhemoglobin at 2.2 A resolution: structure and solvent comparisons of R-state, R2-state and T-state hemoglobins.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
4O51
 
 | | Crystal structure of the QAA variant of anti-hinge rabbit antibody 2095-2 in complex with IDES hinge peptide | | Descriptor: | IDES hinge peptide, QAA-2095-2 heavy chain, QAA-2095-2 light chain | | Authors: | Malia, T.J, Luo, J, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-12-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
