7MQW
 
 | |
6L2B
 
 | |
8HUA
 
 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
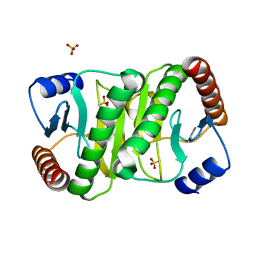
3V1N
 
 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3V1K
 
 | |
3V1M
 
 | | Crystal Structure of the S112A/H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
7O2Y
 
 | |
7O3K
 
 | |
3V1L
 
 | |
7C90
 
 | | Crystal structure of Cytochrome CL from the marine methylotrophic bacterium Methylophaga aminisulfidivorans MPT (Ma-CytcL) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Cytochrome c, ... | | Authors: | Ghosh, S, Dhanasingh, I, Lee, S.H. | | Deposit date: | 2020-06-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of CytochromecLfrom the Aquatic Methylotrophic BacteriumMethylophaga aminisulfidivoransMPT.
J Microbiol Biotechnol., 30, 2020
|
|
4LYE
 
 | |
4LYD
 
 | |
5F1E
 
 | | Apo protein of Sandercyanin | | Descriptor: | Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Ramaswamy, S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EZ2
 
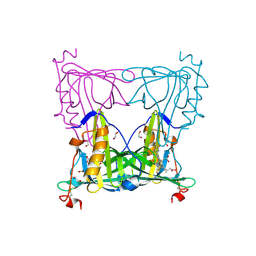 | | Sandercyanin Fluorescent Protein (SFP) | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Ramaswamy, S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F6Z
 
 | | Sandercyanin Fluorescent Protein purified from Sander vitreus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Yu, C.L, Ferraro, D, Sudha, S, Pal, S, Schaefer, W, Gibson, D.T, Subramanian, R. | | Deposit date: | 2015-12-07 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.248 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RAM
 
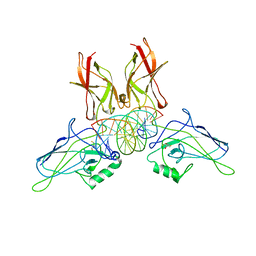 | | A NOVEL DNA RECOGNITION MODE BY NF-KB P65 HOMODIMER | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA (5'-D(*CP*GP*GP*CP*TP*GP*GP*AP*AP*AP*TP*TP*TP*CP*CP*AP*GP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR NF-KB P65) | | Authors: | Chen, Y.-Q, Ghosh, S, Ghosh, G. | | Deposit date: | 1997-11-22 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel DNA recognition mode by the NF-kappa B p65 homodimer.
Nat.Struct.Biol., 5, 1998
|
|
2RAM
 
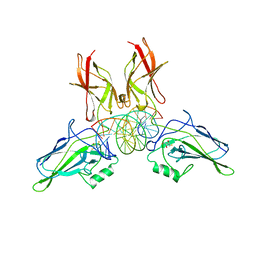 | | A NOVEL DNA RECOGNITION MODE BY NF-KB P65 HOMODIMER | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, DNA (5'-D(*CP*GP*GP*CP*TP*GP*GP*AP*AP*AP*TP*(5IU)P*(5IU)P*CP*CP*AP*GP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR NF-KB P65) | | Authors: | Chen, Y.Q, Ghosh, S, Ghosh, G. | | Deposit date: | 1997-11-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel DNA recognition mode by the NF-kappa B p65 homodimer.
Nat.Struct.Biol., 5, 1998
|
|
1K3Z
 
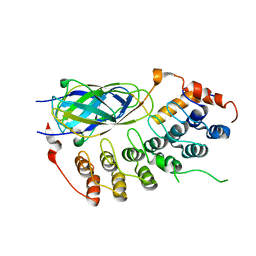 | | X-ray crystal structure of the IkBb/NF-kB p65 homodimer complex | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Shiva, M, Huang, D.B, Chen, Y, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
7YX1
 
 | |
8K65
 
 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase without pump laser irradiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Zoric, D, Sandelin, E, Nango, E, Tanaka, R, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
8K6Y
 
 | | Serial femtosecond crystallography structure of photo dissociated CO from ba3- type cytochrome c oxidase determined by extrapolation method | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Zoric, D, Sandelin, E, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
6EWN
 
 | | HspA from Thermosynechococcus vulcanus in the presence of 2M urea with initial stages of denaturation | | Descriptor: | HspA, UREA | | Authors: | Adir, N, Ghosh, S, Salama, F, Dines, M. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Biophysical and structural characterization of the small heat shock protein HspA from Thermosynechococcus vulcanus in 2 M urea.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
1NFK
 
 | | STRUCTURE OF THE NUCLEAR FACTOR KAPPA-B (NF-KB) P50 HOMODIMER | | Descriptor: | DNA (5'-D(*TP*GP*GP*GP*AP*AP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Ghosh, G, Van Duyne, G, Ghosh, S, Sigler, P.B. | | Deposit date: | 1995-02-28 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of NF-kappa B p50 homodimer bound to a kappa B site.
Nature, 373, 1995
|
|
1OY3
 
 | | CRYSTAL STRUCTURE OF AN IKBBETA/NF-KB P65 HOMODIMER COMPLEX | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Malek, S, Huang, D.B, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
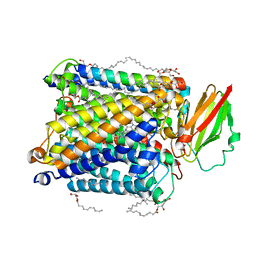
3NV9
 
 | | Crystal structure of Entamoeba histolytica Malic Enzyme | | Descriptor: | ACETATE ION, GLYCEROL, Malic enzyme, ... | | Authors: | Dutta, D, Chakrabarty, A, Ghosh, S.K, Das, A.K. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of Entamoeba histolytica Malic enzyme
To be Published
|
|
