7SMI
 
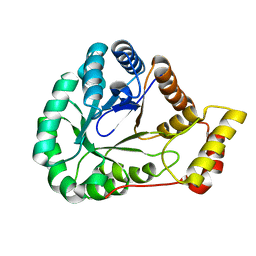 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea | | Descriptor: | L-galactose dehydrogenase | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
4WCH
 
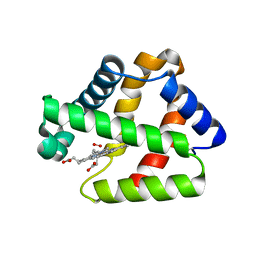 | | Structure of Isolated D Chain of Gigant Hemoglobin from Glossoscolex paulistus | | Descriptor: | Isolated Chain D of Gigant Hemoglobin from Glossoscolex Paulistus, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bachega, J.F.R, Maluf, F.V, Pereira, H.M, Brandao-Neto, J, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-09-04 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8FWP
 
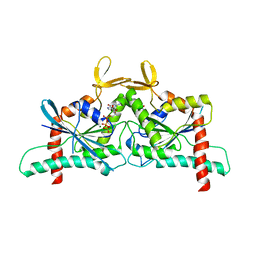 | | Crystal Structure of CDC10 - CDC3 heterocomplex from Saccharomyces cerevisiae | | Descriptor: | Cell division control protein 10, Cell division control protein 3, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Silva, R.M, Leonardo, D.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
J.Struct.Biol., 215, 2023
|
|
8DKT
 
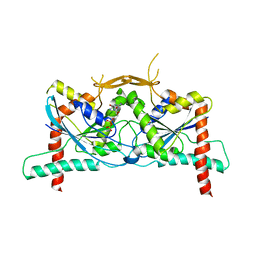 | | Crystal Structure of Septin1 - Septin2 heterocomplex from Drosophila melanogaster | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | de Freitas, A.F, Leonardo, D.A, Cavini, I.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2022-07-06 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conservation and divergence of the G-interfaces of Drosophila melanogaster septins.
Cytoskeleton (Hoboken), 80, 2023
|
|
3CQD
 
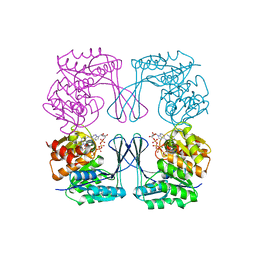 | | Structure of the tetrameric inhibited form of phosphofructokinase-2 from Escherichia coli | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Ambrosio, A.L, Cabrera, R, Caniuguir, A, Garratt, R.C, Babul, J. | | Deposit date: | 2008-04-02 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic structure of phosphofructokinase-2 from Escherichia coli in complex with two ATP molecules. Implications for substrate inhibition.
J.Mol.Biol., 383, 2008
|
|
1XC6
 
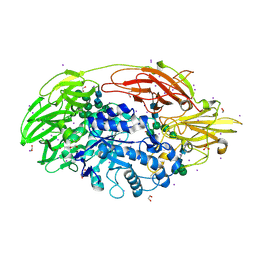 | | Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
5TIR
 
 | | Crystal Structure of Mn Superoxide Dismutase mutant M27V from Trichoderma reesei | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Mendoza, E.R, Brandao-Neto, J, Pereira, H.M, Ferreira Junior, J.R.S, Garratt, R.C. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Mn Superoxide Dismutase mutant M27V from Trichoderma reesei
To Be Published
|
|
3N1C
 
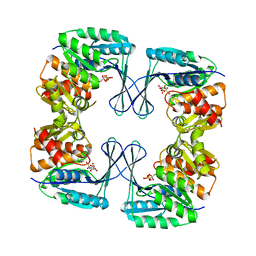 | | Crystal structure of the phosphofructokinase-2 from Escherichia coli in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase isozyme 2 | | Authors: | Pereira, H.M, Cabrera, R, Caniuguir, A, Garratt, R.C, Babul, J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Complex of Phosphofructokinase-2 of Escherichia coli with Fructose-6-phosphate: KINETIC AND STRUCTURAL ANALYSIS OF THE ALLOSTERIC ATP INHIBITION.
J.Biol.Chem., 286, 2011
|
|
1HOT
 
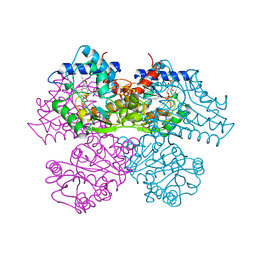 | | GLUCOSAMINE 6-PHOSPHATE DEAMINASE COMPLEXED WITH THE ALLOSTERIC ACTIVATOR N-ACETYL-GLUCOSAMINE-6-PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.L, Garratt, R, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-11-17 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
9BHT
 
 | |
9BHW
 
 | |
7SVQ
 
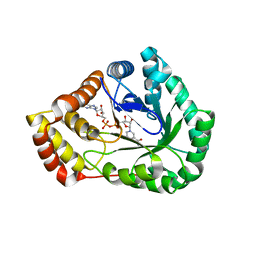 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea in complex with NAD+ | | Descriptor: | L-galactose dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-11-19 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
1HOR
 
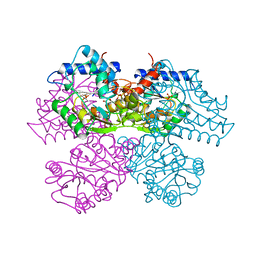 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1DEA
 
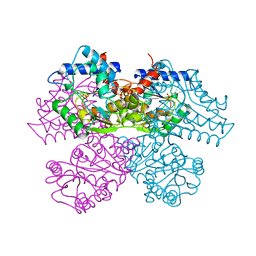 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
8USU
 
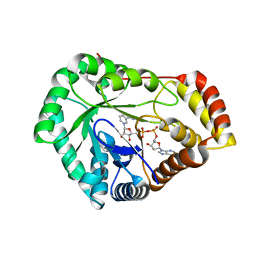 | | Crystal Structure of L-galactose 1-dehydrogenase of Myrciaria dubia in complex with NAD | | Descriptor: | L-galactose dehydrogenase isoform X1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
5KKG
 
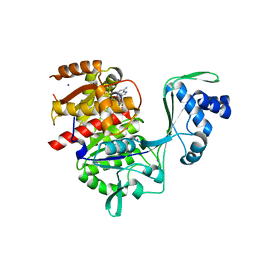 | | Crystal structure of E72A mutant of ancestral protein ancMT of ADP-dependent sugar kinases family | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, IODIDE ION, ... | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-06-21 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
8SGD
 
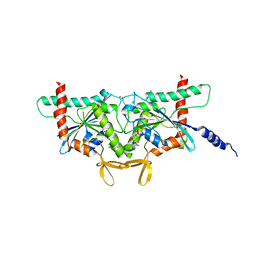 | | Crystal Structure of CDC3(G) - CDC10(Delta 1-10) heterocomplex from Saccharomyces cerevisiae | | Descriptor: | CDC10 isoform 1, CDC3 isoform 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Saladino, G.C.R, Leonardo, D.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
J.Struct.Biol., 215, 2023
|
|
2GO2
 
 | |
5K27
 
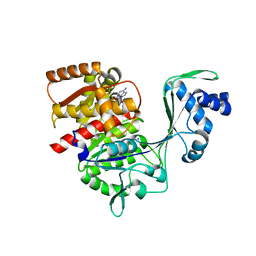 | | Crystal structure of ancestral protein ancMT of ADP-dependent sugar kinases family. | | Descriptor: | ADENOSINE MONOPHOSPHATE, IODIDE ION, ancMT | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
4KVA
 
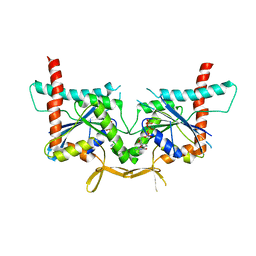 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
4KV9
 
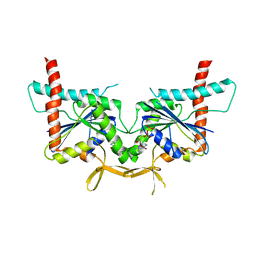 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
7SML
 
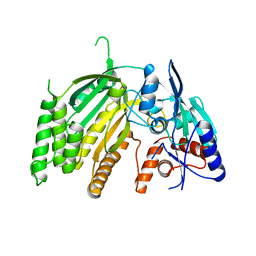 | | Crystal Structure of L-GALACTONO-1,4-LACTONE DEHYDROGENASE de Myrciaria dubia | | Descriptor: | L-GALACTONO-1,4-LACTONE DEHYDROGENASE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2021-10-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit Camu-Camu.
J.Exp.Bot., 2024
|
|
7KSN
 
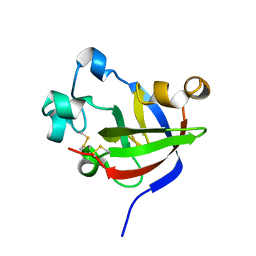 | | Crystal Structure of Sugarwin | | Descriptor: | Sugarwin | | Authors: | Maia, L.B.L, Pereira, H.M, Henrique-Silva, F, Garratt, R.C, Silva Filho, M.C. | | Deposit date: | 2020-11-23 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural and Evolutionary Analyses of PR-4 SUGARWINs Points to a Different Pattern of Protein Function
Frontiers in Plant Science, 12, 2021
|
|
6TFD
 
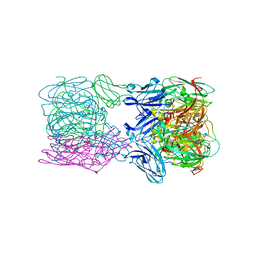 | | Crystal structure of nitrite and NO bound three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
6TFO
 
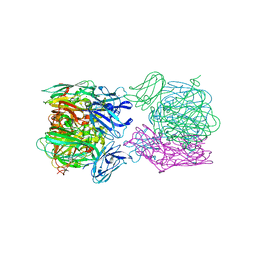 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
