6L1W
 
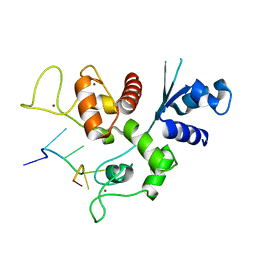 | | Zinc-finger Antiviral Protein (ZAP) bound to RNA | | Descriptor: | RNA (5'-R(*CP*GP*UP*CP*GP*U)-3'), ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Luo, X, Wang, X, Gao, Y, Zhu, J, Liu, S, Gao, G, Gao, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Molecular Mechanism of RNA Recognition by Zinc-Finger Antiviral Protein.
Cell Rep, 30, 2020
|
|
6J9M
 
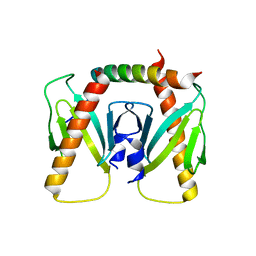 | | NmeBH+AcrIIC2 | | Descriptor: | AcrIIC2, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9K
 
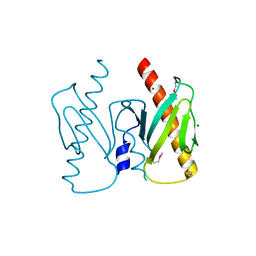 | | Apo-AcrIIC2 | | Descriptor: | AcrIIC2, MAGNESIUM ION | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6J9L
 
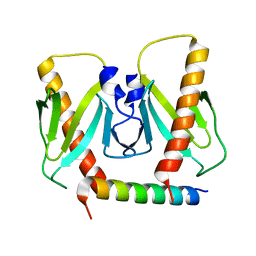 | | FnoBH+AcrIIC2 | | Descriptor: | AcrIIC2, HNH endonuclease family protein | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
6MV1
 
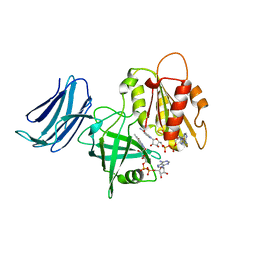 | | 2.15A resolution structure of the CS-b5R domains of human Ncb5or (NAD+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6MV2
 
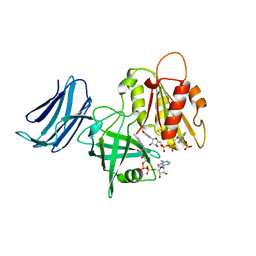 | | 2.05A resolution structure of the CS-b5R domains of human Ncb5or (NADP+ form) | | Descriptor: | Cytochrome b5 reductase 4, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Benson, D.R, Cooper, A, Gao, P, Zhu, H. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the naturally fused CS and cytochrome b5reductase (b5R) domains of Ncb5or reveal an expanded CS fold, extensive CS-b5R interactions and productive binding of the NAD(P)+nicotinamide ring.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6J9N
 
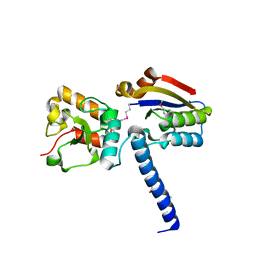 | | NmeHNH+AcrIIC3 | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | Authors: | Zhu, Y.L, Gao, A, Serganov, A, Gao, P. | | Deposit date: | 2019-01-23 | | Release date: | 2019-03-06 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins.
Mol. Cell, 74, 2019
|
|
5ZQ3
 
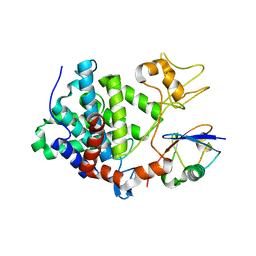 | | PDE-Ubiquitin | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ2
 
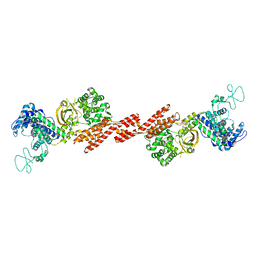 | | SidE apo form | | Descriptor: | SidE | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ7
 
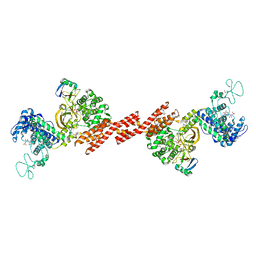 | | SidE-Ubi-NAD | | Descriptor: | ADENOSINE MONOPHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SidE, ... | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ6
 
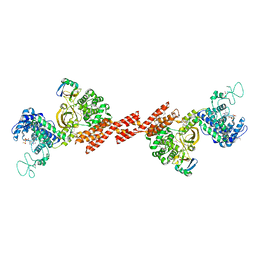 | | SidE-Ubi-ADPr | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ5
 
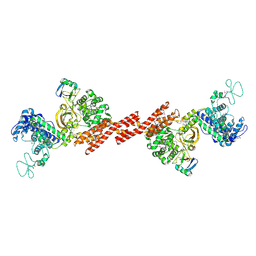 | | SidE-Ubi | | Descriptor: | SidE, Ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
5ZQ4
 
 | | PDE-Ubi-ADPr | | Descriptor: | ADENOSINE MONOPHOSPHATE, SidE, ubiquitin | | Authors: | Wang, Y, Gao, A, Gao, P. | | Deposit date: | 2018-04-17 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE.
Cell, 173, 2018
|
|
3OKG
 
 | | Crystal structure of HsdS subunit from Thermoanaerobacter tengcongensis | | Descriptor: | Restriction endonuclease S subunits, SULFATE ION | | Authors: | Liang, D, Gao, P, Tang, Q, An, X, Yan, X. | | Deposit date: | 2010-08-24 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase
Plos One, 6, 2011
|
|
3GM5
 
 | | Crystal structure of a putative methylmalonyl-coenzyme A epimerase from Thermoanaerobacter tengcongensis at 2.0 A resolution | | Descriptor: | CITRIC ACID, Lactoylglutathione lyase and related lyases, SULFATE ION | | Authors: | Liang, D.-C, Shi, L, Gao, P, Yan, X.-X. | | Deposit date: | 2009-03-12 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative methylmalonyl-coenzyme a epimerase from Thermoanaerobacter tengcongensis at 2.0 A resolution
Proteins, 77, 2009
|
|
5HKW
 
 | | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Apo c-Cbl TKBD Refined to 2.25 A Resolution
To be published
|
|
5HKZ
 
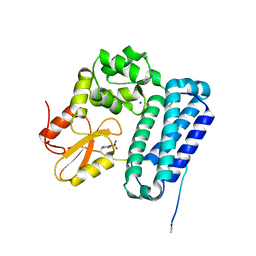 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form) | | Descriptor: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2, SODIUM ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8 A Resolution (P21 form)
To be published
|
|
5HKX
 
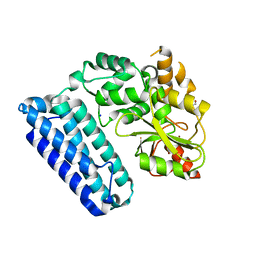 | | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CBL, SODIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of c-Cbl TKBD-RING domains (Y371E mutant) Refined to 1.85 A Resolution
To be published
|
|
5HL0
 
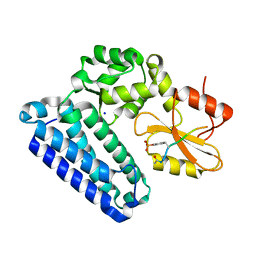 | | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution | | Descriptor: | E3 ubiquitin-protein ligase CBL, SODIUM ION, Sprouty 2 (SPRY2) | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of c-Cbl TKBD in complex with SPRY2 peptide (54-60, pY55) Refined to 2.2A Resolution
To Be Published
|
|
5HKY
 
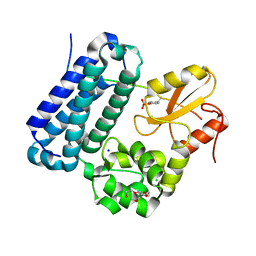 | | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase CBL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Zhang, N, Cooper, A, Gao, P, Perez, R.P. | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of c-Cbl TKBD domain in complex with SPRY2 peptide (36-60, pY55) Refined to 1.8A Resolution (P6 form)
To be published
|
|
8WKW
 
 | | Structure of MAVS-CARD Filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Shi, M, Gao, P. | | Deposit date: | 2023-09-28 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of MAVS-CARD Filament
To Be Published
|
|
8IL7
 
 | |
8WX7
 
 | | Crystal structure of SHP2 in complex with JAB-3186 | | Descriptor: | (5~{S})-1'-[6-azanyl-5-(2-azanyl-3-chloranyl-pyridin-4-yl)sulfanyl-pyrazin-2-yl]spiro[5,7-dihydrocyclopenta[b]pyridine-6,4'-piperidine]-5-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Ma, C, Gao, P, Kang, D, Han, H, Sun, X, Zhang, W, Qian, D, Wang, Y, Long, W. | | Deposit date: | 2023-10-27 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of JAB-3312, a Potent SHP2 Allosteric Inhibitor for Cancer Treatment.
J.Med.Chem., 67, 2024
|
|
5XZE
 
 | | Mouse cGAS bound to the inhibitor RU332 | | Descriptor: | (3R)-3-[1-(3H-1lambda~4~,3-benzothiazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5YBB
 
 | | Structural basis underlying complex assembly andconformational transition of the type I R-M system | | Descriptor: | DNA, Restriction endonuclease S subunits, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, Y.P, Tang, Q, Zhang, J.Z, Tian, L.F, Gao, P, Yan, X.X. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-29 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis underlying complex assembly and conformational transition of the type I R-M system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
