4O2C
 
 | | An Nt-acetylated peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
1VGK
 
 | | The crystal structure of class I Major histocompatibility complex, H-2Kd at 2.0 A resolution | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Zhou, M, Xu, Y, Liu, Y, Gao, G.F, Tien, P, Rao, Z. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The crystal structure of class I major histocompatibility complex, H-2Kd at 2.0 A resolution
To be Published
|
|
4MX1
 
 | | Structure of ricin A chain bound with 2-amino-4-oxo-N-(2-(3-phenylureido)ethyl)-3,4-dihydropteridine-7-carboxamide | | Descriptor: | 2-amino-4-oxo-N-{2-[(phenylcarbamoyl)amino]ethyl}-3,4-dihydropteridine-7-carboxamide, MALONIC ACID, Ricin A chain, ... | | Authors: | Robertus, J.D, Wiget, P.A, Manzano, L.A, Pruet, J.M, Gao, G, Saito, R, Jasheway, K.R, Monzingo, A.F, Anslyn, E.V. | | Deposit date: | 2013-09-25 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sulfur incorporation generally improves Ricin inhibition in pterin-appended glycine-phenylalanine dipeptide mimics.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LLA
 
 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
5WZ2
 
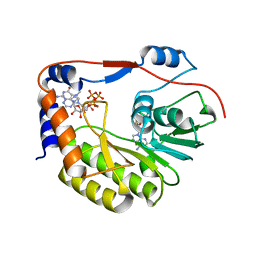 | | Crystal structure of Zika virus NS5 methyltransferase bound to SAM and RNA analogue (m7GpppA) | | Descriptor: | NS5 MTase, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE | | Authors: | Duan, W, Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of Zika virus NS5 reveals conserved drug targets.
EMBO J., 36, 2017
|
|
5XEB
 
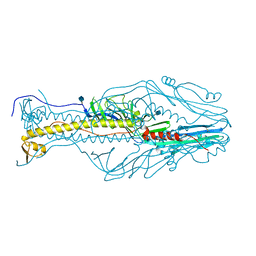 | | Structure of the envelope glycoprotein of Dhori virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Peng, R, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-04-03 | | Release date: | 2017-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structures of human-infectingThogotovirusfusogens support a common ancestor with insect baculovirus
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XL7
 
 | |
5WWJ
 
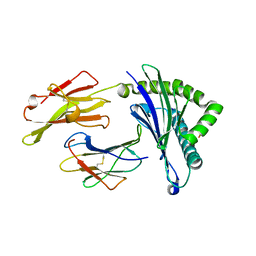 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1).
To Be Published
|
|
5XL4
 
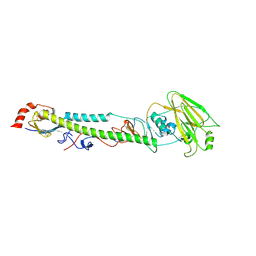 | |
5XLA
 
 | |
5XL8
 
 | |
5XLB
 
 | |
4HZX
 
 | | Crystal structure of influenza A neuraminidase N3 complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZZ
 
 | | Crystal structure of influenza neuraminidase N3-H274Y complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4HZV
 
 | | The crystal structure of influenza A neuraminidase N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4I00
 
 | | Crystal structure of influenza A neuraminidase N3-H274Y complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
4F7T
 
 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PB1 (498-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
5GSR
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5HGA
 
 | | HLA*A2402 complex with HIV nef138 Y2F-8mer mutant epitope | | Descriptor: | 8-mer from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
3U9G
 
 | | Crystal structure of the Zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Chen, S, Xu, Y, Zhang, K, Wang, X, Sun, J, Gao, G, Liu, Y. | | Deposit date: | 2011-10-18 | | Release date: | 2012-03-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5HGD
 
 | | HLA*A2402 complexed with HIV nef138 Y2F mutant 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
3V32
 
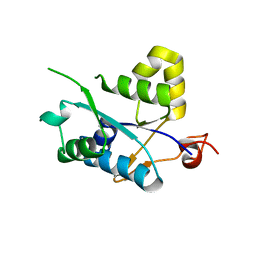 | | Crystal structure of MCPIP1 N-terminal conserved domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
4H52
 
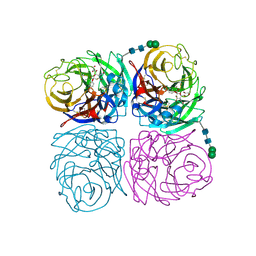 | | Wild-type influenza N2 neuraminidase covalent complex with 3-fluoro-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
8KC2
 
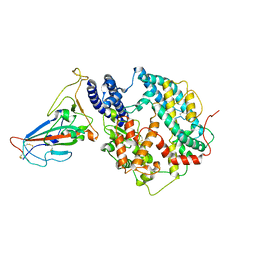 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
