3ICN
 
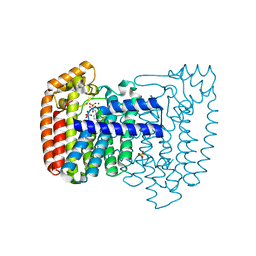 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with isopentenyl pyrophosphate and 3-Fluoro-1-(2-hydroxy-2,2-bis-phosphono-ethyl)-pyridinium | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-17 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
6WW1
 
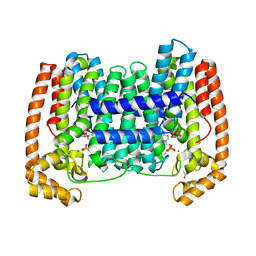 | | Crystal structure of the LmFPPS mutant E97Y | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Gabelli, S.B. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
6NCK
 
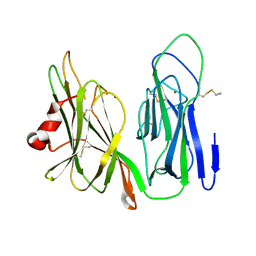 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, NICKEL (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Miller, M.S, Maheshwari, S, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6NCT
 
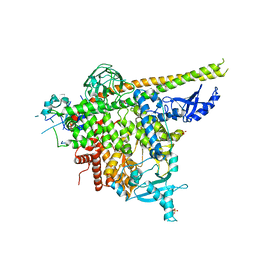 | | Structure of p110alpha/niSH2 - vector data collection | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, SULFATE ION, ... | | Authors: | Miller, M.S, Maheshwari, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
6O9C
 
 | |
3ICM
 
 | | Trypanosoma cruzi farnesyl diphosphate synthase homodimer in complex with isopentenyl pyrophosphate, Mg2+ and 1-(2-Hydroxy-2,2-bis-phosphono-ethyl)-3-phenyl-pyridinium | | Descriptor: | 1-(2-hydroxy-2,2-diphosphonoethyl)-3-phenylpyridinium, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Amzel, L.M, Huang, C.H, Gabelli, S.B, Oldfield, E. | | Deposit date: | 2009-07-17 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of nitrogen-containing bisphosphonates (N-BPs) to the Trypanosoma cruzi farnesyl diphosphate synthase homodimer.
Proteins, 78, 2010
|
|
3NPE
 
 | | Structure of VP14 in complex with oxygen | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-cis-epoxycarotenoid dioxygenase 1, chloroplastic, ... | | Authors: | Messing, S.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-06-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid.
Plant Cell, 22, 2010
|
|
6W51
 
 | |
6AMP
 
 | | Crystal structure of H172A PHM (CuH absent, CuM present) | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6ALV
 
 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AY0
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) soaked with peptide | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AO6
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-15 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AN3
 
 | | Crystal structure of H172A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant soaked with peptide (no CuH bound, no peptide bound) | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
6ALA
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5VY2
 
 | | Crystal structure of the F36A mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-05-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
5W6X
 
 | |
5WJA
 
 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WKW
 
 | | Crystal structure of apo wild type peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-07-25 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
5WM0
 
 | |
1MP2
 
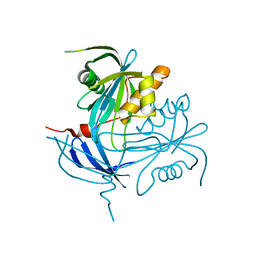 | | Structure of MT-ADPRase (Apoenzyme), a Nudix hydrolase from Mycobacterium tuberculosis | | Descriptor: | ADPR pyrophosphatase | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MQE
 
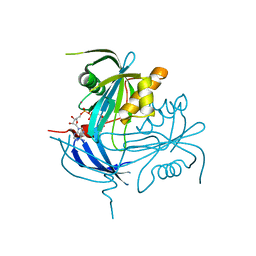 | | Structure of the MT-ADPRase in complex with gadolidium and ADP-ribose, a Nudix enzyme | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADPR pyrophosphatase, GADOLINIUM ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MK1
 
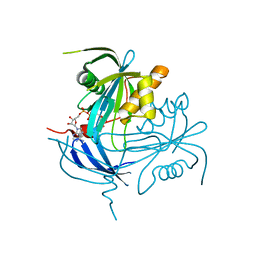 | | Structure of the MT-ADPRase in complex with ADPR, a Nudix enzyme | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADPR pyrophosphatase | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-08-28 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MR2
 
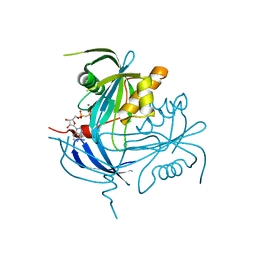 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | Descriptor: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MQW
 
 | | Structure of the MT-ADPRase in complex with three Mn2+ ions and AMPCPR, a Nudix enzyme | | Descriptor: | ADPR pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MANGANESE (II) ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
