4LQ4
 
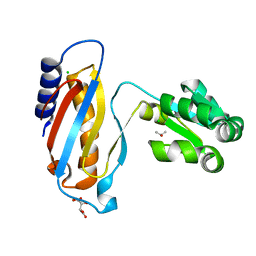 | | crystal structure of mutant ribosomal protein L1 from Methanococcus jannaschii with deletion of 8 residues from C-terminus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, S.V, Shkliaeva, A.A, Garber, M.B, Nikonov, S.V, Sarskikh, A.V. | | Deposit date: | 2013-07-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a mutant of archaeal ribosomal protein L1 from Methanococcus jannaschii
Crystallography Reports, 59, 2014
|
|
8AIP
 
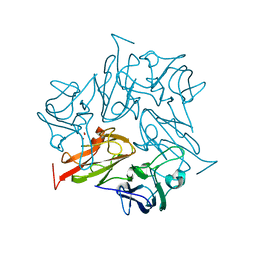 | | Crystal Structure of Two-domain bacterial laccase from the actinobacterium Streptomyces carpinensis VKM Ac-1300 | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, Two-Domain Laccase | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Trubitsina, L, Trubitsin, I, Leontievsky, A, Lisov, A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Novel Two-Domain Laccase with Middle Redox Potential: Physicochemical and Structural Properties.
Biochemistry Mosc., 88, 2023
|
|
5ML7
 
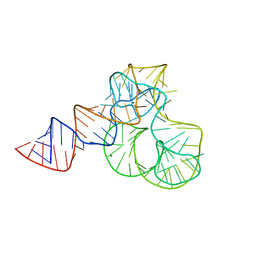 | | CRYSTAL STRUCTURE OF L1-STALK FRAGMENT OF 23S rRNA FROM HALOARCULA MARISMORTUI | | Descriptor: | 23S ribosomal RNA, CALCIUM ION, MAGNESIUM ION | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the 23S rRNA Fragment Specific to r-Protein L1 and Designed Model of the Ribosomal L1 Stalk from Haloarcula marismortui
Crystals, 2017
|
|
5NPM
 
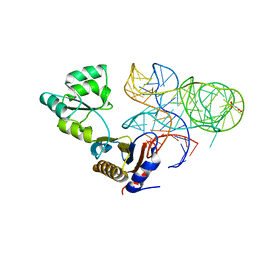 | | CRYSTAL STRUCTURE OF MUTANT RIBOSOMAL PROTEIN TTHL1 LACKING 8 N-TERMINAL RESIDUES IN COMPLEX WITH 80NT 23S RNA FROM THERMUS THERMOPHILUS | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, MALONATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Nevskaya, N.A, Nikonov, S.V, Garber, M.B. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | [Influence of Nonconserved Regions of L1 Protuberance of Thermus thermophilus Ribosome on the Affinity of L1 Protein to 23s rRNA].
Mol.Biol.(Moscow), 52, 2018
|
|
5O4I
 
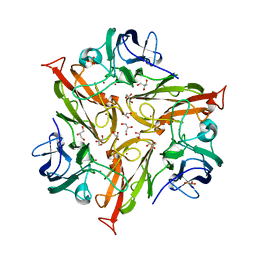 | |
5O3K
 
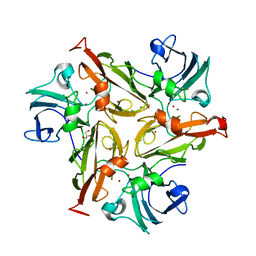 | |
5O4Q
 
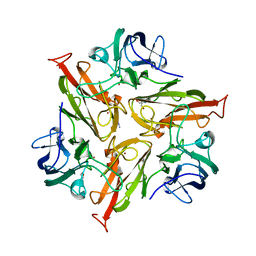 | |
6Q53
 
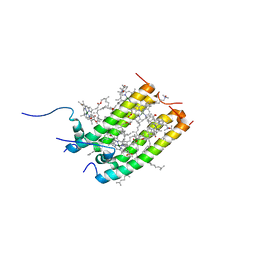 | |
6Z27
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LCP crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
6Z02
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV in surfo crystallization | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Fufina, T.Y, Vasilieva, L.G, Betzel, C. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
6Z1J
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP co-crystallization with spheroidene | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
8QKY
 
 | | Bacteriophage T5 dUTPase | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, GLYCEROL | | Authors: | Gabdulkhakov, A.G, Dzhus, U.F, Selikhanov, G.K, Glukhov, A.S. | | Deposit date: | 2023-09-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacteriophage T5 dUTPase: Combination of Common Enzymatic and Novel Functions.
Int J Mol Sci, 25, 2024
|
|
8QLD
 
 | | Bacteriophage T5 dUTPase mutant with loop deletion (30-35 aa) | | Descriptor: | CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Dzhus, U.F, Selikhanov, G.K, Glukhov, A.S. | | Deposit date: | 2023-09-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacteriophage T5 dUTPase: Combination of Common Enzymatic and Novel Functions.
Int J Mol Sci, 25, 2024
|
|
8C3F
 
 | | Double mutant I(L177)H/F(M197)H structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2022-12-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Properties and Crystal Structure of the Cereibacter sphaeroides Photosynthetic Reaction Center with Double Amino Acid Substitution I(L177)H + F(M197)H.
Membranes (Basel), 13, 2023
|
|
8AF1
 
 | | Beta-Lytic Protease from Lysobacter capsici | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kudryakova, I.V, Afoshin, A.S, Vasilyeva, N.V. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Int J Mol Sci, 23, 2022
|
|
6RHQ
 
 | | Crystal Structure of Two-Domain Laccase mutant I170A from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6S0O
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces griseoflavus produced at 0.25 mM copper sulfate in growth medium | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-06-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
6RH9
 
 | | Crystal Structure of Two-Domain Laccase mutant I170F from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-04-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
7P0Q
 
 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (100K, 26keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-Ray structure of Rhodobacter sphaeroides reaction center with M197 Phe-His substitution clarifies properties of the mutant complex
To Be Published
|
|
7P17
 
 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 12keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-07-01 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | X-Ray structure of Rhodobacter sphaeroides reaction center with M197 Phe-His substitution clarifies properties of the mutant complex
To Be Published
|
|
7P2C
 
 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 26keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-07-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray structure of the Rhodobacter sphaeroides reaction center with an M197 Phe→His substitution clarifies the properties of the mutant complex.
Iucrj, 9, 2022
|
|
4RJL
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kravchenko, O.V, Nikonov, O.S, Arhipova, V.I, Stolboushkina, E.A, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2014-10-09 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6401 Å) | | Cite: | Crystal structure of gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in complex with GDPCP at 1.64A resolution
To be Published
|
|
4XR5
 
 | | X-ray structure of the unliganded thymidine phosphorylase from Salmonella typhimurium at 2.05 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YEK
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YYY
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with uridine | | Descriptor: | CITRIC ACID, TRIETHYLENE GLYCOL, Thymidine phosphorylase, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-03-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
