8KCE
 
 | |
5B1Y
 
 | | Crystal structure of NADPH bound carbonyl reductase from Aeropyrum pernix | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, Y, Araki, T, Ohshima, T. | | Deposit date: | 2015-12-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Catalytic properties and crystal structure of thermostable NAD(P)H-dependent carbonyl reductase from the hyperthermophilic archaeon Aeropyrum pernix K1.
Enzyme.Microb.Technol., 91, 2016
|
|
7BXX
 
 | | Tetanus neurotoxin translocation domain -C467S | | Descriptor: | Tetanus toxin | | Authors: | Zhang, C.M, Imoto, Y, Fukuda, Y, Yamashita, E, Inoue, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | SOLUTION SCATTERING (2.34 Å), X-RAY DIFFRACTION | | Cite: | Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X, 5, 2021
|
|
7BY5
 
 | | Tetanus neurotoxin mutant-(H233A/E234Q/H237A/Y375F) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Zhang, C.M, Imoto, Y, Fukuda, Y, Yamashita, E, Inoue, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural flexibility of the tetanus neurotoxin revealed by crystallographic and solution scattering analyses.
J Struct Biol X, 5, 2021
|
|
6LZX
 
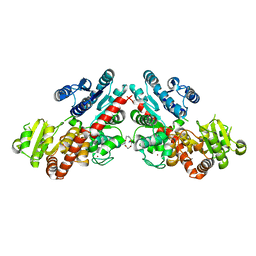 | | Structure of Phytolacca americana UGT3 with 15-crown-5 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13-pentaoxacyclopentadecane, BROMIDE ION, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Crown-ether-mediated crystal structures of the glycosyltransferase PaGT3 from Phytolacca americana.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LZY
 
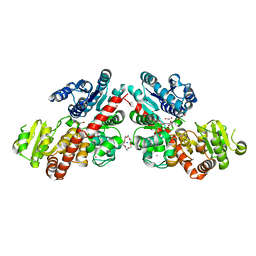 | | Structure of Phytolacca americana UGT3 with 18-crown-6 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, BROMIDE ION, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2020-02-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crown-ether-mediated crystal structures of the glycosyltransferase PaGT3 from Phytolacca americana.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5YXW
 
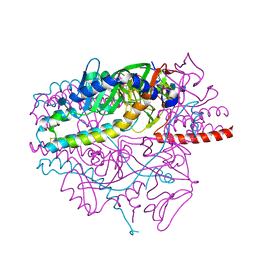 | | Crystal structure of the prefusion form of measles virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-07 | | Release date: | 2018-02-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZD
 
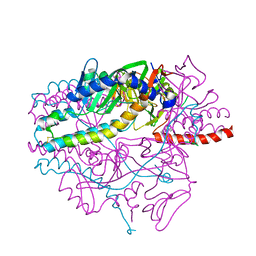 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor peptide (FIP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZC
 
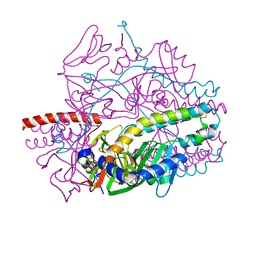 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor compound (AS-48) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitro-2-[(phenylacetyl)amino]benzamide, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7VEL
 
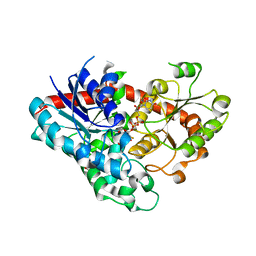 | | Crystal structure of Phytolacca americana UGT3 with UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VEJ
 
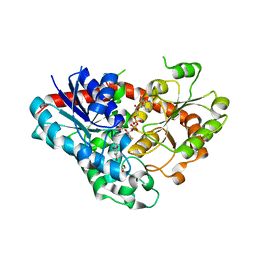 | | Crystal structure of Phytolacca americana UGT3 with kaempferol and UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VEK
 
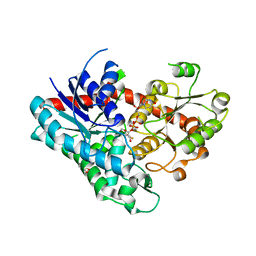 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6JEN
 
 | | Structure of Phytolacca americana UGT2 complexed with UDP-2fluoro-glucose and pterostilbene | | Descriptor: | Glycosyltransferase, Pterostilbene, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2019-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An Ambidextrous Polyphenol GlycosyltransferasePaGT2 fromPhytolacca americana.
Biochemistry, 59, 2020
|
|
6JEL
 
 | | Structure of Phytolacca americana apo UGT2 | | Descriptor: | Glycosyltransferase | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2019-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Ambidextrous Polyphenol GlycosyltransferasePaGT2 fromPhytolacca americana.
Biochemistry, 59, 2020
|
|
6JEM
 
 | | Structure of Phytolacca americana UGT2 complexed with UDP-2fluoro-glucose and resveratrol | | Descriptor: | Glycosyltransferase, RESVERATROL, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2019-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Ambidextrous Polyphenol GlycosyltransferasePaGT2 fromPhytolacca americana.
Biochemistry, 59, 2020
|
|
5ZIQ
 
 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 4 | | Descriptor: | 1,2-ETHANEDIOL, Globin protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-03-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
3WE7
 
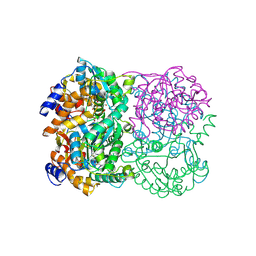 | | Crystal Structure of Diacetylchitobiose Deacetylase from Pyrococcus horikoshii | | Descriptor: | ACETIC ACID, GLYCEROL, HEXANE-1,6-DIOL, ... | | Authors: | Mine, S, Nakamura, T, Fukuda, Y, Inoue, T, Uegaki, K, Sato, T. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-07 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
5ZM9
 
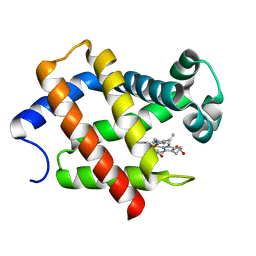 | | Crystal structure of hexacoordinated heme protein from anhydrobiotic tardigrade at pH 7 | | Descriptor: | CHLORIDE ION, Globin Protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kim, J, Fukuda, Y, Inoue, T. | | Deposit date: | 2018-04-02 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Kumaglobin: a hexacoordinated heme protein from an anhydrobiotic tardigrade, Ramazzottius varieornatus.
FEBS J., 286, 2019
|
|
7YPP
 
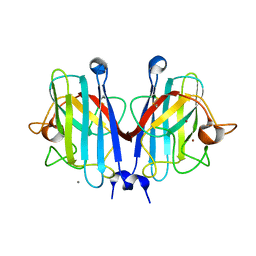 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Sim, K.-S, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7YPR
 
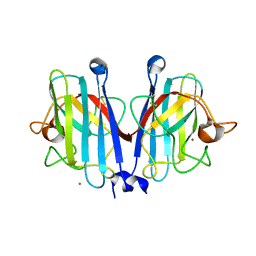 | | Structural basis of a superoxide dismutase from a tardigrade, Ramazzottius varieornatus strain YOKOZUNA-1. | | Descriptor: | COPPER (II) ION, POTASSIUM ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Sim, K.-S, Fukuda, Y, Inoue, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of a superoxide dismutase from a tardigrade: Ramazzottius varieornatus strain YOKOZUNA-1.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3WXB
 
 | | Crystal structure of NADPH bound carbonyl reductase from chicken fatty liver | | Descriptor: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein | | Authors: | Yoneda, K, Sakuraba, H, Fukuda, Y, Sone, T, Araki, T, Ohshima, T. | | Deposit date: | 2014-07-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel NAD(P)H-dependent carbonyl reductase specifically expressed in the thyroidectomized chicken fatty liver: catalytic properties and crystal structure.
Febs J., 282, 2015
|
|
