1ZOX
 
 | |
2HG0
 
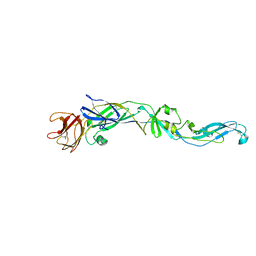 | | Structure of the West Nile Virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | Authors: | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
5KVG
 
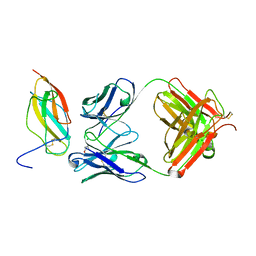 | | Zika specific antibody, ZV-67, bound to ZIKA envelope DIII | | Descriptor: | CHLORIDE ION, ZIKA Envelope DIII, ZV-67 Antibody Fab Heavy Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
5KVF
 
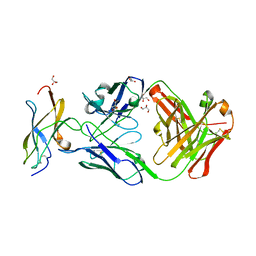 | | Zika specific antibody, ZV-64, bound to ZIKA envelope DIII | | Descriptor: | GLYCEROL, ZV-64 Antibody Fab Heavy Chain, ZV-64 Antibody Fab Light Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
2NZ1
 
 | |
5KVE
 
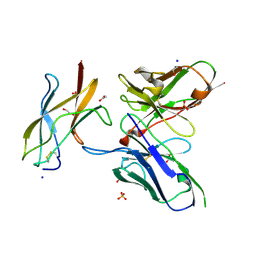 | | Zika specific antibody, ZV-48, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Genome polyprotein, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
5KVD
 
 | | Zika specific antibody, ZV-2, bound to ZIKA envelope DIII | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SODIUM ION, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
2NYZ
 
 | |
2GRK
 
 | |
2FWO
 
 | |
6PLK
 
 | |
7K9H
 
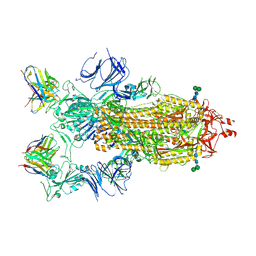 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7K9J
 
 | |
6N08
 
 | |
6MYM
 
 | |
4PDC
 
 | |
6MXU
 
 | |
6MZK
 
 | |
7K9K
 
 | |
4OIG
 
 | |
4OII
 
 | |
7K9I
 
 | |
1XAK
 
 | |
4OIE
 
 | |
1F3J
 
 | | HISTOCOMPATIBILITY ANTIGEN I-AG7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, LYSOZYME C, ... | | Authors: | Latek, R.R, Unanue, E.R, Fremont, D.H. | | Deposit date: | 2000-06-04 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice.
Immunity, 12, 2000
|
|
