3I5T
 
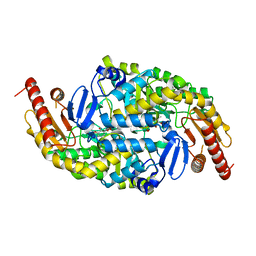 | | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides KD131 | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides
To be Published
|
|
3I4S
 
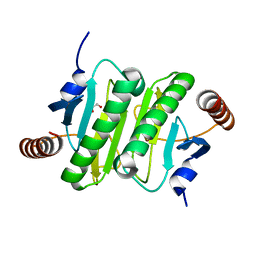 | | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN blr8122 FROM Bradyrhizobium japonicum | | Descriptor: | GLYCEROL, HISTIDINE TRIAD PROTEIN | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN FROM Bradyrhizobium japonicum
To be Published
|
|
3GKB
 
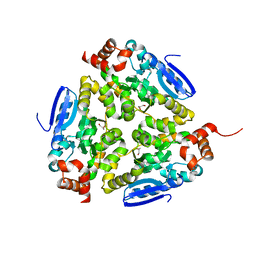 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3H7A
 
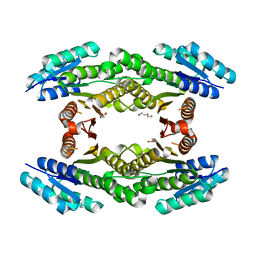 | | CRYSTAL STRUCTURE OF SHORT-CHAIN DEHYDROGENASE FROM Rhodopseudomonas palustris | | Descriptor: | UNKNOWN LIGAND, short chain dehydrogenase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CRYSTAL STRUCTURE OF SHORT-CHAIN DEHYDROGENASE FROM Rhodopseudomonas palustris
To be Published
|
|
3IGH
 
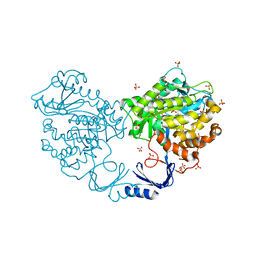 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus horikoshii ot3 | | Descriptor: | SULFATE ION, UNCHARACTERIZED METAL-DEPENDENT HYDROLASE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Horikoshii
To be Published
|
|
3K9D
 
 | | CRYSTAL STRUCTURE OF PROBABLE ALDEHYDE DEHYDROGENASE FROM Listeria monocytogenes EGD-e | | Descriptor: | ALDEHYDE DEHYDROGENASE, CHLORIDE ION, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase from Listeria Monocytogenes
To be Published
|
|
3PVE
 
 | | Crystal structure of the G2 domain of Agrin from Mus Musculus | | Descriptor: | Agrin, Agrin protein | | Authors: | Sampathkumar, P, Do, J, Bain, K, Freeman, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the G2 domain of Agrin from Mus Musculus
To be Published
|
|
3GMF
 
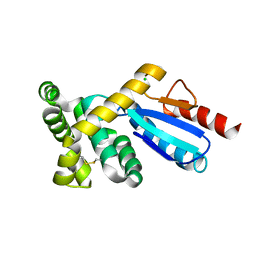 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3HCW
 
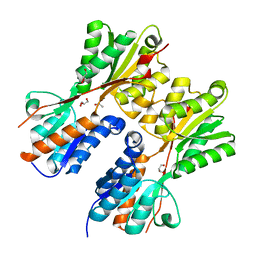 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | Descriptor: | GLYCEROL, Maltose operon transcriptional repressor | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
3IA1
 
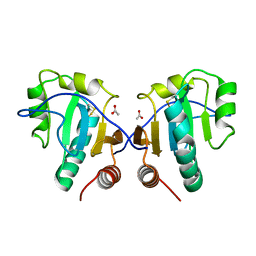 | | Crystal structure of thio-disulfide isomerase from Thermus thermophilus | | Descriptor: | ACETATE ION, Thio-disulfide isomerase/thioredoxin | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of thio-disulfide isomerase from Thermus thermophilus
To be Published
|
|
3IBM
 
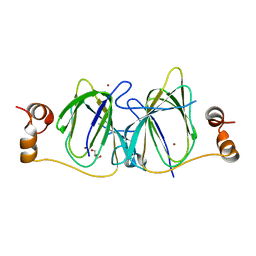 | | CRYSTAL STRUCTURE OF cupin 2 domain-containing protein Hhal_0468 FROM Halorhodospira halophila | | Descriptor: | Cupin 2, conserved barrel domain protein, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-16 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF cupin 2 domain-containing PROTEIN Hhal_0468 FROM Halorhodospira halophila
To be Published
|
|
3JU2
 
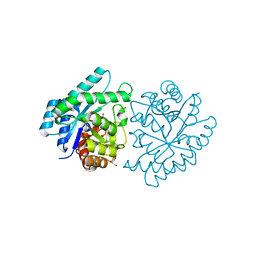 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
3HV2
 
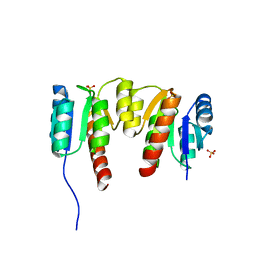 | | Crystal structure of signal receiver domain OF HD domain-containing protein FROM Pseudomonas fluorescens Pf-5 | | Descriptor: | Response regulator/HD domain protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of signal receiver domain oF HD domain-containing protein 3 FROM Pseudomonas fluorescens
To be Published
|
|
3GT7
 
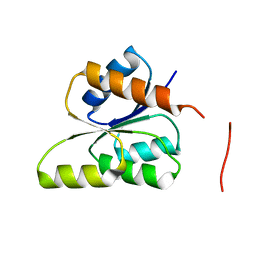 | | CRYSTAL STRUCTURE OF SIGNAL RECEIVER DOMAIN OF SIGNAL TRANSDUCTION HISTIDINE KINASE FROM Syntrophus aciditrophicus | | Descriptor: | Sensor protein | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Signal Transduction Kinase from Syntrophus Aciditrophicus
To be Published
|
|
2IRM
 
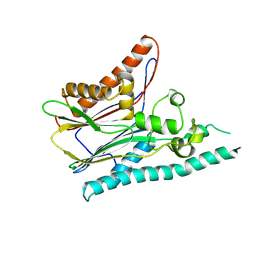 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2IQ1
 
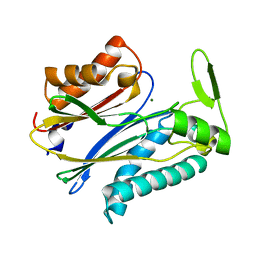 | | Crystal structure of human PPM1K | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C kappa, PPM1K | | Authors: | Bonanno, J.B, Freeman, J, Russell, M, Bain, K.T, Adams, J, Pelletier, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2HY3
 
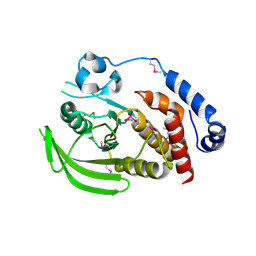 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3F13
 
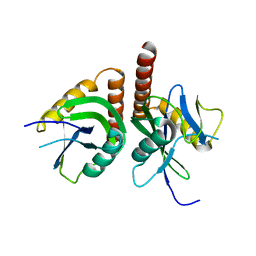 | | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum | | Descriptor: | putative nudix hydrolase family member | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative nudix hydrolase family member from Chromobacterium violaceum
To be Published
|
|
3CYJ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3DTD
 
 | | Crystal structure of invasion associated protein b from bartonella henselae | | Descriptor: | GLYCEROL, Invasion-associated protein B | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Slocombe, A, Groshong, C, Koss, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Invasion Associated Protein B from Bartonella Henselae.
To be Published
|
|
2FH7
 
 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3DBI
 
 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
3DBY
 
 | | Crystal structure of uncharacterized protein from Bacillus cereus G9241 (CSAP Target) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, uncharacterized protein | | Authors: | Ramagopal, U.A, Bonanno, J.B, Ozyurt, S, Freeman, J, Wasserman, S, Hu, S, Groshong, C, Rodgers, L, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of uncharacterized protein from Bacillus cereus G9241
To be published
|
|
3E61
 
 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
3E3M
 
 | | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi | | Descriptor: | Transcriptional regulator, LacI family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-07 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a LacI family transcriptional regulator from Silicibacter pomeroyi
To be Published
|
|
