4XV5
 
 | | CcP gateless cavity | | Descriptor: | BENZIMIDAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV7
 
 | | CcP gateless cavity | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV8
 
 | | CcP gateless cavity | | Descriptor: | BENZAMIDINE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV6
 
 | | CcP gateless cavity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4XV4
 
 | | CcP gateless cavity | | Descriptor: | 2-AMINO-5-METHYLTHIAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
7S1I
 
 | | Wild-type Escherichia coli stalled ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1J
 
 | | Wild-type Escherichia coli ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2RDH
 
 | | Crystal structure of Staphylococcal Superantigen-Like protein 11 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Superantigen-like protein 11 | | Authors: | Chung, M.C, Wines, B.D, Baker, H, Langley, R.J, Baker, E.N, Fraser, J.D. | | Deposit date: | 2007-09-24 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition
Mol.Microbiol., 66, 2007
|
|
7FR6
 
 | |
7FR5
 
 | |
7FRA
 
 | |
7FR2
 
 | |
7FR7
 
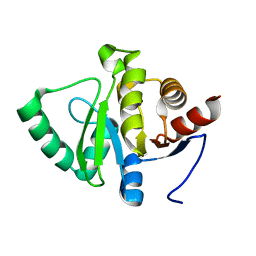 | |
7FR0
 
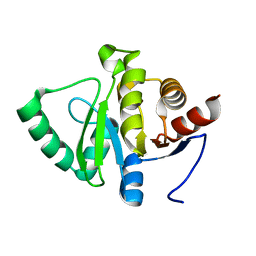 | |
7FR1
 
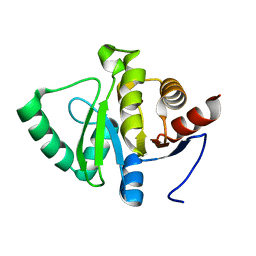 | |
7FR8
 
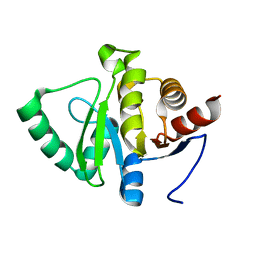 | |
7FRB
 
 | |
7FR4
 
 | |
7FR3
 
 | |
7FRC
 
 | |
7FRD
 
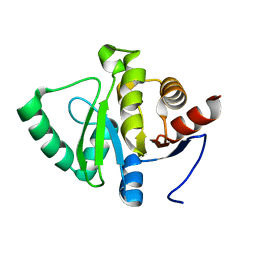 | |
7FR9
 
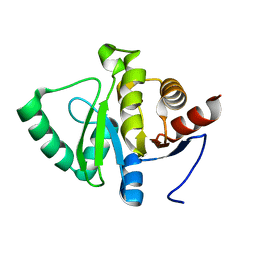 | |
3KLS
 
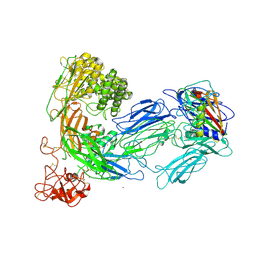 | | Structure of complement C5 in complex with SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KM9
 
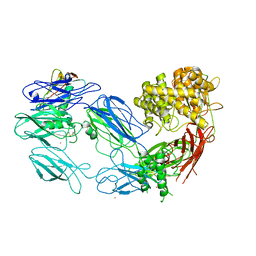 | | Structure of complement C5 in complex with the C-terminal beta-grasp domain of SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4OPR
 
 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying G238S mutation | | Descriptor: | CALCIUM ION, SULFATE ION, TEM-94 ES-beta-lactamase, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-06 | | Release date: | 2015-05-20 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
