9HNP
 
 | |
4W6V
 
 | | Crystal structure of a peptide transporter from Yersinia enterocolitica at 3 A resolution | | Descriptor: | Di-/tripeptide transporter | | Authors: | Jeckelmann, J.-M, Boggavarapu, R, Harder, D, Ucurum, Z, Fotiadis, D. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.01819 Å) | | Cite: | Role of electrostatic interactions for ligand recognition and specificity of peptide transporters.
Bmc Biol., 13, 2015
|
|
8CNK
 
 | |
8CQD
 
 | |
8CQC
 
 | |
7ZNY
 
 | | Cryo-EM structure of the canine distemper virus tetrameric attachment glycoprotein | | Descriptor: | Hemagglutinin glycoprotein | | Authors: | Kalbermatter, D, Jeckelmann, J.-M, Wyss, M, Plattet, P, Fotiadis, D. | | Deposit date: | 2022-04-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and supramolecular organization of the canine distemper virus attachment glycoprotein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7B03
 
 | |
7O82
 
 | | The L-arginine/agmatine antiporter from E. coli at 1.7 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine/agmatine antiporter, DECANE, ... | | Authors: | Jeckelmann, J.M, Ilgue, H, Kalbermatter, D, Fotiadis, D. | | Deposit date: | 2021-04-14 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structure of the amino acid transporter AdiC reveals insights into the role of water molecules and networks in oligomerization and substrate binding.
Bmc Biol., 19, 2021
|
|
7Q0L
 
 | | Crystal structure of the peptide transporter YePEPT-K314A at 2.93 A | | Descriptor: | Peptide transporter YePEPT | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Boggavarapu, R, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
7Q0M
 
 | | Crystal structure of the peptide transporter YePEPT-K314A in complex with LZNV at 2.66 A | | Descriptor: | (2~{S})-2-[[(2~{S})-2-azanyl-6-[(4-nitrophenyl)methoxycarbonylamino]hexanoyl]amino]-3-methyl-butanoic acid, Peptide transporter YePEPT, UNDECYL-MALTOSIDE | | Authors: | Jeckelmann, J.M, Stauffer, M, Ilgue, H, Fotiadis, D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Peptide transporter structure reveals binding and action mechanism of a potent PEPT1 and PEPT2 inhibitor.
Commun Chem, 5, 2022
|
|
8QSR
 
 | |
8QST
 
 | |
8A6L
 
 | |
6G9X
 
 | | Crystal structure of a MFS transporter at 2.54 Angstroem resolution | | Descriptor: | 2-sulfanylbenzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MERCURY (II) ION, ... | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic basis of L-lactate transport in the SLC16 solute carrier family.
Nat Commun, 10, 2019
|
|
6HCL
 
 | | Crystal structure of a MFS transporter with Ligand at 2.69 Angstroem resolution | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Major facilitator superfamily MFS_1, nonyl beta-D-glucopyranoside | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2018-08-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of L-lactate transport in the SLC16 solute carrier family.
Nat Commun, 10, 2019
|
|
5J4I
 
 | |
5J4N
 
 | |
6XYE
 
 | |
6ZGR
 
 | | Crystal structure of a MFS transporter with bound 1-hydroxynaphthalene-2-carboxylic acid at 2.67 Angstroem resolution | | Descriptor: | 1-hydroxynaphthalene-2-carboxylic acid, L-lactate transporter | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
6ZGU
 
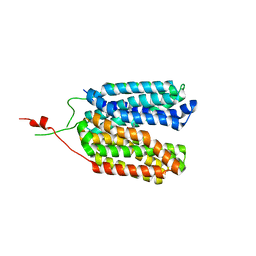 | | Crystal structure of a MFS transporter with bound 3-(2-methylphenyl)propanoic acid at 2.41 Angstroem resolution | | Descriptor: | 3-(2-methylphenyl)propanoic acid, L-lactate transporter | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
6ZGT
 
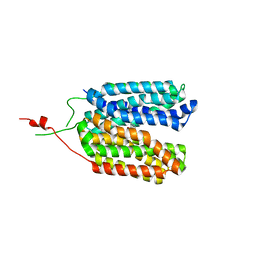 | | Crystal structure of a MFS transporter with bound 2-naphthoic acid at 2.39 Angstroem resolution | | Descriptor: | L-lactate transporter, naphthalene-2-carboxylic acid | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
6ZGS
 
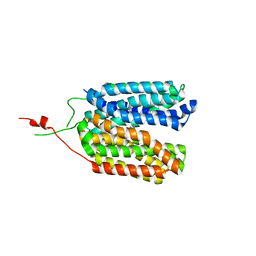 | | Crystal structure of a MFS transporter with bound 3-phenylpropanoic acid at 2.39 Angstroem resolution | | Descriptor: | HYDROCINNAMIC ACID, L-lactate transporter | | Authors: | Kalbermatter, D, Bosshart, P, Bonetti, S, Fotiadis, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | The making of a potent L-lactate transport inhibitor
Commun Chem, 4, 2021
|
|
