4DCY
 
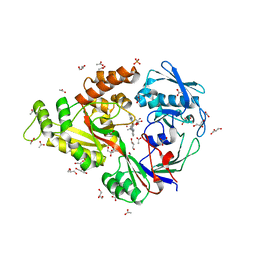 | | X-ray structure of NikA in complex with Fe(1S,2S)-N,N-kappa-Bis(2-pyridylmethyl)-N-carboxymethyl-N-kappa-methyl-1,2-cyclohexanediamine | | Descriptor: | ACETATE ION, GLYCEROL, Nickel-binding periplasmic protein, ... | | Authors: | Cherrier, M.V, Girgenti, E, Amara, P, Iannello, M, Marchi-Delapierre, C, Fontecilla-Camps, J.C, Menage, S, Cavazza, C. | | Deposit date: | 2012-01-18 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the periplasmic nickel-binding protein NikA provides insights for artificial metalloenzyme design.
J.Biol.Inorg.Chem., 17, 2012
|
|
4P3X
 
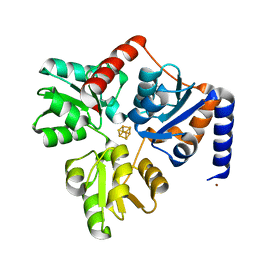 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
4R5F
 
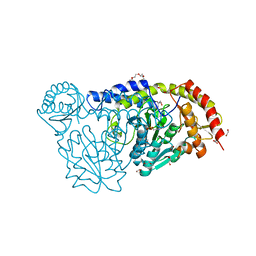 | | X-ray structure of the D199K mutant of the cysteine desulfurase IscS from A. fulgidus | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETATE ION, CALCIUM ION, ... | | Authors: | Pagnier, A, Nicolet, Y, Fontecilla-Camps, J.C. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IscS from Archaeoglobus fulgidus has no desulfurase activity but may provide a cysteine ligand for [Fe2S2] cluster assembly.
Biochim.Biophys.Acta, 1853, 2015
|
|
5CVR
 
 | |
5E44
 
 | | Crystal structure of holo-FNR of A. fischeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium.
Sci Adv, 1, 2015
|
|
2Q60
 
 | | Crystal structure of the ligand binding domain of polyandrocarpa misakiensis rxr in tetramer in absence of ligand | | Descriptor: | Retinoid X receptor | | Authors: | Borel, F, De Groot, A, Juillan-Binard, C, De Rosny, E, Laudet, V, Pebay-Peyroula, E, Fontecilla-Camps, J.-C, Ferrer, J.-L. | | Deposit date: | 2007-06-04 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ligand-binding domain of the retinoid X receptor from the ascidian polyandrocarpa misakiensis.
Proteins, 74, 2008
|
|
2UZA
 
 | | CRYSTAL STRUCTURE OF THE FREE RADICAL INTERMEDIATE OF PYRUVATE:FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Cavazza, C, Contreras-Martel, C, Fontecilla-Camps, J.C. | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Flexibility of thiamine diphosphate revealed by kinetic crystallographic studies of the reaction of pyruvate-ferredoxin oxidoreductase with pyruvate.
Structure, 14, 2006
|
|
4GD3
 
 | | Structure of E. coli hydrogenase-1 in complex with cytochrome b | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the O(2)-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b.
Structure, 21, 2013
|
|
5N08
 
 | |
5N07
 
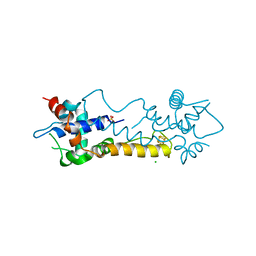 | |
2FRV
 
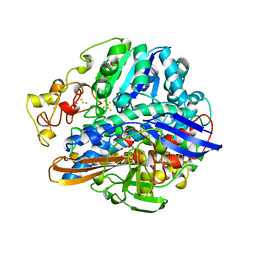 | | CRYSTAL STRUCTURE OF THE OXIDIZED FORM OF NI-FE HYDROGENASE | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Frey, M, Fontecilla-Camps, J.C. | | Deposit date: | 1997-06-10 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structure of the [Nife] Hydrogenase Active Site: Evidence for Biologically Uncommon Fe Ligands
J.Am.Chem.Soc., 118, 1996
|
|
4UPV
 
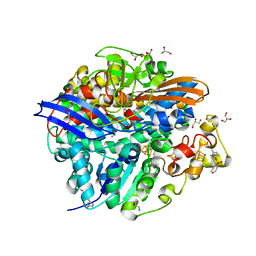 | | Low X-ray dose structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-18 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4UPE
 
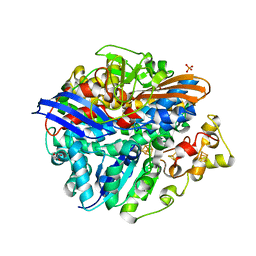 | | Structure of the unready Ni-A state of the S499C mutant of D. fructosovorans NiFe-hydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
1XIU
 
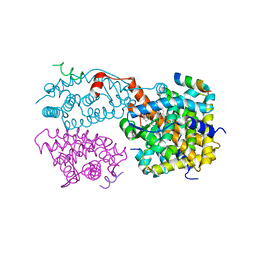 | | Crystal structure of the agonist-bound ligand-binding domain of Biomphalaria glabrata RXR | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 1, RXR-like protein | | Authors: | De Groot, A, De Rosny, E, Juillan-Binard, C, Ferrer, J.-L, Laudet, V, Pebay-Peroula, E, Fontecilla-Camps, J.-C, Borel, F. | | Deposit date: | 2004-09-22 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Novel Tetrameric Complex of Agonist-bound Ligand-binding Domain of Biomphalaria glabrata Retinoid X Receptor.
J.Mol.Biol., 354, 2005
|
|
3IIZ
 
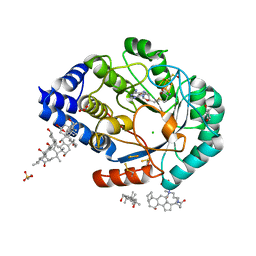 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with S-adenosyl-L-methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Biotin synthetase, putative, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
3IIX
 
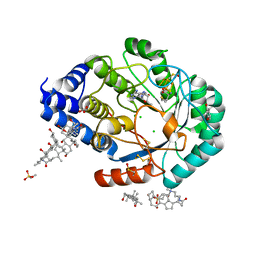 | | X-ray structure of the FeFe-hydrogenase maturase HydE from T. maritima in complex with methionine and 5'deoxyadenosine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CARBONATE ION, ... | | Authors: | Nicolet, Y, Amara, P, Mouesca, J.M, Fontecilla-Camps, J.C. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Unexpected electron transfer mechanism upon AdoMet cleavage in radical SAM proteins
Proc.Natl.Acad.Sci.USA, 2009
|
|
1FDT
 
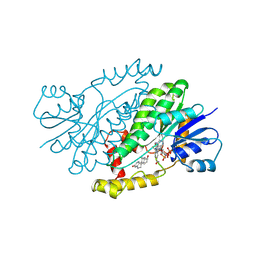 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Housset, D, Breton, R, Mazza, C, Fontecilla-Camps, J.-C. | | Deposit date: | 1996-06-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex of human 17beta-hydroxysteroid dehydrogenase with estradiol and NADP+ identifies two principal targets for the design of inhibitors.
Structure, 4, 1996
|
|
3UQY
 
 | | H2-reduced structure of E. coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USE
 
 | | Crystal Structure of E. coli hydrogenase-1 in its as-isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USC
 
 | | Crystal Structure of E. coli hydrogenase-1 in a ferricyanide-oxidized form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GIT
 
 | | Crystal structure of a truncated acetyl-CoA synthase | | Descriptor: | Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Volbeda, A, Darnault, C, Fontecilla-Camps, J.C. | | Deposit date: | 2009-03-06 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel domain arrangement in the crystal structure of a truncated acetyl-CoA synthase from Moorella thermoacetica
Biochemistry, 48, 2009
|
|
4R33
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan and S-adenosyl-L-homocysteine bound | | Descriptor: | CHLORIDE ION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4R34
 
 | | X-ray structure of the tryptophan lyase NosL with Tryptophan, 5'-deoxyadenosine and methionine bound | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Nicolet, Y, Zeppieri, L, Amara, P, Fontecilla-Camps, J.-C. | | Deposit date: | 2014-08-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Tryptophan Lyase (NosL): Evidence for Radical Formation at the Amino Group of Tryptophan.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3CIW
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FeFe-Hydrogenase maturase, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
6H8T
 
 | | Crystal structure of Papain modify by achiral Ru(II)complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Papain, ... | | Authors: | Cherrier, M.V, Amara, P, Talbi, B, Salmin, M, Fontecilla-Camps, J.C. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic evidence for unexpected selective tyrosine hydroxylations in an aerated achiral Ru-papain conjugate.
Metallomics, 10, 2018
|
|
