1ZCO
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphoheptonate aldolase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Schofield, L.R, Anderson, B.F, Patchett, M.L, Norris, G.E, Jameson, G.B, Parker, E.J. | | Deposit date: | 2005-04-12 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Ambiguity and Crystal Structure of Pyrococcus furiosus 3-Deoxy-d-arabino-heptulosonate-7-phosphate Synthase: An Ancestral 3-Deoxyald-2-ulosonate-phosphate Synthase?(,)
Biochemistry, 44, 2005
|
|
5L32
 
 | |
3R1P
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, form P1 | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1V
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges, in complex with an azo compound | | Descriptor: | 4-{(E)-[4-(propan-2-yl)phenyl]diazenyl}phenol, Odorant binding protein, antennal | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
3R1O
 
 | | Odorant Binding Protein 7 from Anopheles gambiae with Four Disulfide Bridges | | Descriptor: | Odorant binding protein, antennal, PALMITIC ACID | | Authors: | Lagarde, A, Spinelli, S, Tegoni, M, Field, L, He, X, Zhou, J.J, Cambillau, C. | | Deposit date: | 2011-03-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Odorant Binding Protein 7 from Anopheles gambiae Exhibits an Outstanding Adaptability of Its Binding Site.
J.Mol.Biol., 414, 2011
|
|
2WCJ
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (10E,12Z)-tetradecadien-1-ol | | Descriptor: | (10E,12Z)-tetradeca-10,12-dien-1-ol, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCH
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykal | | Descriptor: | (10E,12Z)-hexadeca-10,12-dienal, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WC5
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, HEXADECA-10,12-DIEN-1-OL, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCM
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (10E)-hexadecen-12-yn-1-ol | | Descriptor: | (10E)-hexadec-10-en-12-yn-1-ol, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCK
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) without ligand | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WCL
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (8E, 10Z)-hexadecadien-1-ol | | Descriptor: | (8E,10Z)-HEXADECA-8,10-DIEN-1-OL, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
2WC6
 
 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with bombykol and water to Arg 110 | | Descriptor: | GENERAL ODORANT-BINDING PROTEIN 1, HEXADECA-10,12-DIEN-1-OL, MAGNESIUM ION | | Authors: | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | Deposit date: | 2009-03-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
4Z45
 
 | | Structure of OBP3 from the currant-lettuce aphid Nasonovia ribisnigri | | Descriptor: | Odorant-binding protein NribOBP3 | | Authors: | Northey, T, Venthur, H, De Biasio, F, Chauviac, F.-X, Cole, A.R, Field, L.M, Zhou, J.-J, Keep, N.H. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structures and Binding Dynamics of Odorant-Binding Protein 3 from two aphid species Megoura viciae and Nasonovia ribisnigri.
Sci Rep, 6, 2016
|
|
4Z39
 
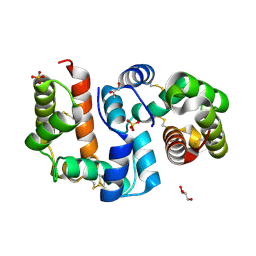 | | Structure of OBP3 from the vetch aphid Megoura viciae | | Descriptor: | GLYCEROL, Odorant-binding protein, SULFATE ION | | Authors: | Northey, T, Venthur, H, De Biasio, F, Chauviac, F.-X, Cole, A.R, Field, L.M, Zhou, J.-J, Keep, N.H. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures and Binding Dynamics of Odorant-Binding Protein 3 from two aphid species Megoura viciae and Nasonovia ribisnigri.
Sci Rep, 6, 2016
|
|
6P8C
 
 | | 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (MthRED) from Methanothermobacter thermautotrophicus | | Descriptor: | 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5'-phosphate reductase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carbone, V, Schofield, L.R, Hannus, I, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-06-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Crystal Structure of 2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (MthRED) from Methanothermobacter thermautotrophicus
To Be Published
|
|
2LPB
 
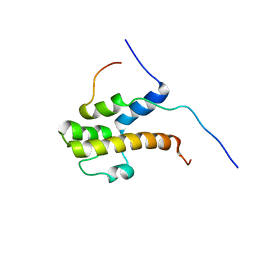 | | Structure of the complex of the central activation domain of Gcn4 bound to the mediator co-activator domain 1 of Gal11/med15 | | Descriptor: | General control protein GCN4, Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Brzovic, P.S, Heikaus, C.C, Kisselev, L, Vernon, R, Herbig, E, Pacheco, D, Warfield, L, Littlefield, P, Baker, D, Klevit, R.E, Hahn, S. | | Deposit date: | 2012-02-07 | | Release date: | 2012-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex.
Mol.Cell, 44, 2011
|
|
2LZE
 
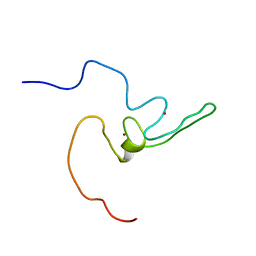 | | Ligase 10C | | Descriptor: | ZINC ION, a primordial catalytic fold generated by in vitro evolution | | Authors: | Chao, F, Morelli, A, Haugner, J, Churchfield, L, Hagmann, L, Shi, L, Masterson, L, Sarangi, R, Veglia, G, Seelig, B. | | Deposit date: | 2012-10-01 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of a primordial catalytic fold generated by in vitro evolution.
Nat.Chem.Biol., 9, 2013
|
|
4RFL
 
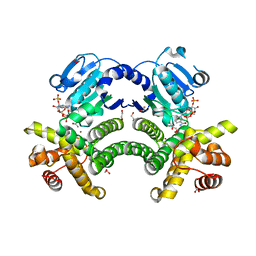 | | Crystal structure of G1PDH with NADPH from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, Glycerol-1-phosphate dehydrogenase [NAD(P)+], NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
4RGQ
 
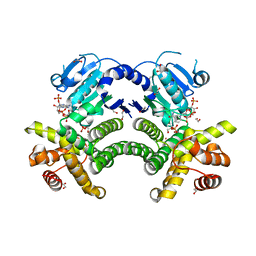 | | Crystal structure of the Methanocaldococcus jannaschii G1PDH with NADPH and DHAP | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, Glycerol-1-phosphate dehydrogenase, ... | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
5L31
 
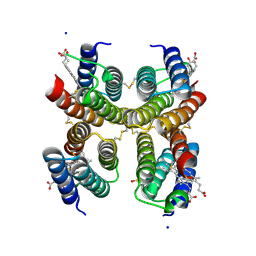 | |
3ZOK
 
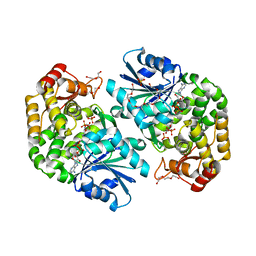 | | Structure of 3-Dehydroquinate Synthase from Actinidia chinensis in complex with NAD | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, DI(HYDROXYETHYL)ETHER, GLYCINE, ... | | Authors: | Mittelstaedt, G, Negron, L, Schofield, L.R, Marsh, K, Parker, E.J. | | Deposit date: | 2013-02-22 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical and structural characterisation of dehydroquinate synthase from the New Zealand kiwifruit Actinidia chinensis.
Arch. Biochem. Biophys., 537, 2013
|
|
4RGV
 
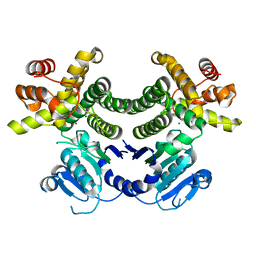 | | Crystal structure of the Methanocaldococcus jannaschii G1PDH | | Descriptor: | Glycerol-1-phosphate dehydrogenase, MAGNESIUM ION, ZINC ION | | Authors: | Carbone, V, Ronimus, R.S, Schofield, L.R, Sutherland-Smith, A.J. | | Deposit date: | 2014-09-30 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Evolution of the Archaeal Lipid Synthesis Enzyme sn-Glycerol-1-phosphate Dehydrogenase.
J.Biol.Chem., 290, 2015
|
|
6ALY
 
 | | Solution structure of yeast Med15 ABD2 residues 277-368 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Tuttle, L.M, Pacheco, D, Warfield, L, Hahn, S, Klevit, R.E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex.
Cell Rep, 22, 2018
|
|
4C1K
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOHEPTONATE ALDOLASE, CADMIUM ION, CARBONATE ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Destabilization of the Homotetrameric Assembly of 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase from the Hyperthermophile Pyrococcus Furiosus Enhances Enzymatic Activity
J.Mol.Biol., 426, 2014
|
|
4C1L
 
 | | Crystal structure of pyrococcus furiosus 3-deoxy-D-arabino- heptulosonate 7-phosphate synthase I181D interface mutant | | Descriptor: | 2-dehydro-3-deoxyphosphoheptonate aldolase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nazmi, A.R, Schofield, L.R, Dobson, R.C.J, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the homotetrameric assembly of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile Pyrococcus furiosus enhances enzymatic activity.
J. Mol. Biol., 426, 2014
|
|
