1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
2X0U
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO A 2-amino substituted benzothiazole scaffold | | Descriptor: | 6,7-DIHYDRO[1,4]DIOXINO[2,3-F][1,3]BENZOTHIAZOL-2-AMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Kaar, J.L, Basse, N, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
2X0V
 
 | | STRUCTURE OF THE P53 CORE DOMAIN MUTANT Y220C BOUND TO 4-(trifluoromethyl)benzene-1,2-diamine | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Basse, N, Kaar, J.L, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2009-12-17 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the Rational Design of P53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant.
Chem.Biol., 17, 2010
|
|
5LAP
 
 | | p53 cancer mutant Y220C with Cys182 alkylation | | Descriptor: | 5-chloranylpyrimidine-4-carboxylic acid, Cellular tumor antigen p53, ZINC ION | | Authors: | Joerger, A.C, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 2-Sulfonylpyrimidines: Mild alkylating agents with anticancer activity toward p53-compromised cells.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2YBG
 
 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XWR
 
 | |
2WXC
 
 | | The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. | | Descriptor: | DIHYDROLIPOYLTRANSSUCCINASE | | Authors: | Neuweiler, H, Sharpe, T.D, Rutherford, T.J, Johnson, C.M, Allen, M.D, Ferguson, N, Fersht, A.R. | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Folding Mechanism of Bbl: Plasticity of Transition-State Structure Observed within an Ultrafast Folding Protein Family.
J.Mol.Biol., 390, 2009
|
|
2L9V
 
 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
2NNT
 
 | | General structural motifs of amyloid protofilaments | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Ferguson, N, Becker, J, Tidow, H, Tremmel, S, Sharpe, T.D, Krause, G, Flinders, J, Petrovich, M, Berriman, J, Oschkinat, H, Fersht, A.R. | | Deposit date: | 2006-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | SOLID-STATE NMR | | Cite: | General structural motifs of amyloid protofilaments.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
1UOL
 
 | |
5AOJ
 
 | | Structure of the p53 cancer mutant Y220C in complex with 2-hydroxy-3, 5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid | | Descriptor: | 2-hydroxy-3,5-diiodo-4-(1H-pyrrol-1-yl)benzoic acid, CELLULAR TUMOR ANTIGEN P53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Joerger, A.C, Baud, M.G, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2015-09-10 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant P53.
Structure, 23, 2015
|
|
2BIO
 
 | |
2BIP
 
 | |
2BIN
 
 | |
2BIM
 
 | |
2BIQ
 
 | |
1SS1
 
 | | STAPHYLOCOCCAL PROTEIN A, B-DOMAIN, Y15W MUTANT, NMR, 25 STRUCTURES | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | Sato, S, Religa, T.L, Daggett, V, Fersht, A.R. | | Deposit date: | 2004-03-23 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From The Cover: Testing protein-folding simulations by experiment: B domain of protein A.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1JON
 
 | |
1HL2
 
 | |
1KID
 
 | |
1KCQ
 
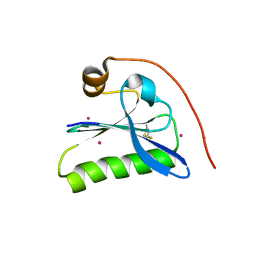 | | Human Gelsolin Domain 2 with a Cd2+ bound | | Descriptor: | CADMIUM ION, GELSOLIN | | Authors: | Kazmirski, S.L, Isaacson, R.L, An, C, Buckle, A, Johnson, C.M, Daggett, V, Fersht, A.R. | | Deposit date: | 2001-11-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Loss of a metal-binding site in gelsolin leads to familial amyloidosis-Finnish type.
Nat.Struct.Biol., 9, 2002
|
|
5NP0
 
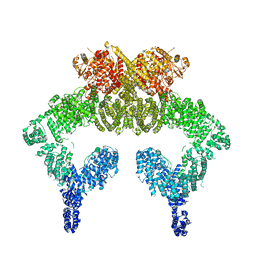 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NP1
 
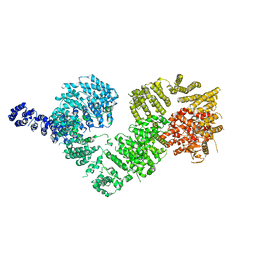 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
2C6A
 
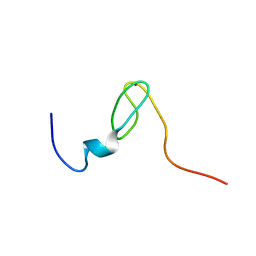 | | Solution structure of the C4 zinc-finger domain of HDM2 | | Descriptor: | UBIQUITIN-PROTEIN LIGASE E3 MDM2, ZINC ION | | Authors: | Yu, G.W, Allen, M.D, Andreeva, A, Fersht, A.R, Bycroft, M. | | Deposit date: | 2005-11-08 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C4 Zinc Finger Domain of Hdm2.
Protein Sci., 15, 2006
|
|
