8YX9
 
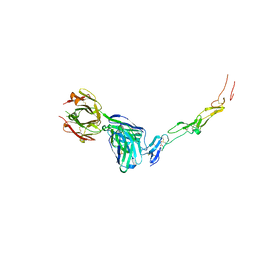 | | CD40 in complex with Dacetuzumab Fab | | Descriptor: | Dacetuzumab, Heavy chain, light chain, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2024-04-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human CD40 in complex with monoclonal antibodies dacetuzumab and bleselumab.
Biochem.Biophys.Res.Commun., 714, 2024
|
|
8YX1
 
 | | CD40 in complex with Bleselumab Fab | | Descriptor: | Bleselumab, heavy chain, light chain, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2024-04-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human CD40 in complex with monoclonal antibodies dacetuzumab and bleselumab.
Biochem.Biophys.Res.Commun., 714, 2024
|
|
8XF7
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XEU
 
 | |
8XFA
 
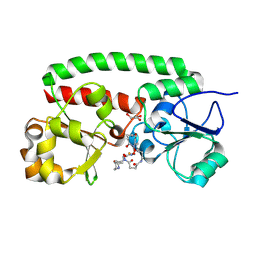 | | Structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes with ferrioxamine E bound | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, FE (III) ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XFI
 
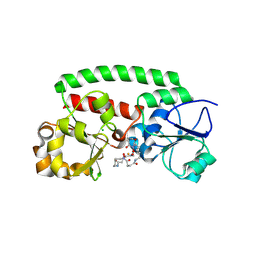 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine E bound (crystal form 2) | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, CHLORIDE ION, FE (III) ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XET
 
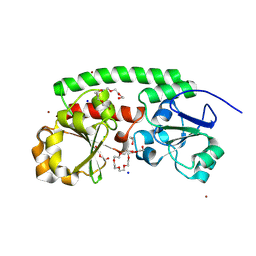 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF4
 
 | | Structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with Bisucaberin bound | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Iron-hydroxamate ABC transporter substrate-binding protein FtsB | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF8
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB from Streptococcus pyogenes with ferrioxamine B | | Descriptor: | 1,2-ETHANEDIOL, Ferrioxamine B, Iron-hydroxamate ABC transporter substrate-binding protein FtsB, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
8XF9
 
 | | High-resolution structure of the siderophore periplasmic binding protein FtsB mutant Y137A from Streptococcus pyogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caaveiro, J.M.M, Fernandez-Perez, J, Tsumoto, K. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis for the ligand promiscuity of the hydroxamate siderophore binding protein FtsB from Streptococcus pyogenes.
Structure, 2024
|
|
