6EZ8
 
 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
6EPC
 
 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
8QXV
 
 | | In situ structure average of GroEL14-GroES7 complexes with narrow GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXS
 
 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with wide GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXU
 
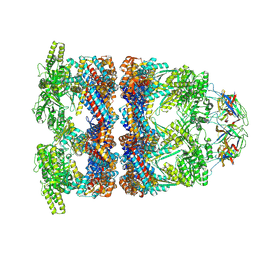 | | In situ structure average of GroEL14-GroES7 complexes with wide GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXT
 
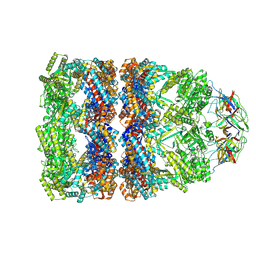 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with narrow GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
7DXJ
 
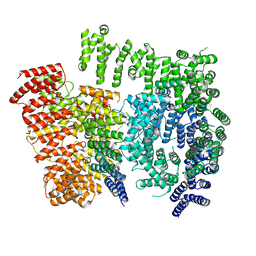 | |
7DXK
 
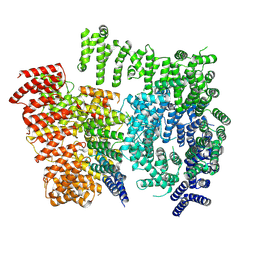 | |
8P4R
 
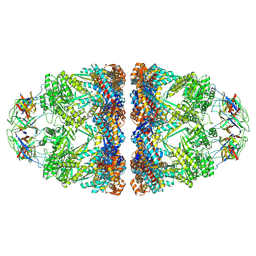 | | In situ structure average of GroEL14-GroES14 complexes in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4M
 
 | | CryoEM structure of a C7-symmetrical GroEL7-GroES7 cage in presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4O
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4N
 
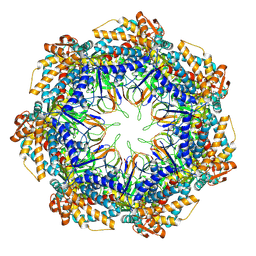 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated disordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4P
 
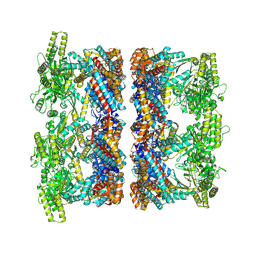 | | Structure average of GroEL14 complexes found in the cytosol of Escherichia coli overexpressing GroEL obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
