4KBX
 
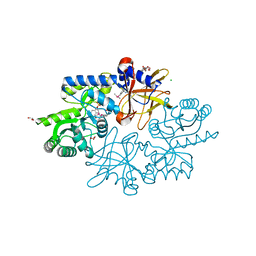 | | Crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx from escherichia coli | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-04-24 | | Release date: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx from escherichia coli
To be Published
|
|
4HCD
 
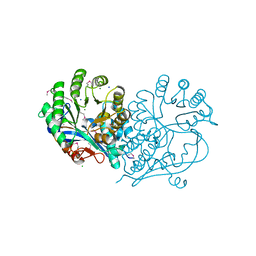 | | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium | | Descriptor: | CHLORIDE ION, Isomerase/lactonizing enzyme, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Millikin, C, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from agrobacterium tumefaciens complexed with magnesium
To be Published
|
|
4LC8
 
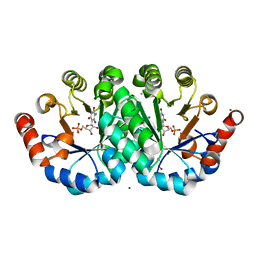 | | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
1X8M
 
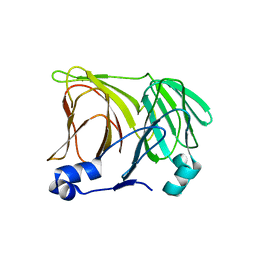 | |
2IM5
 
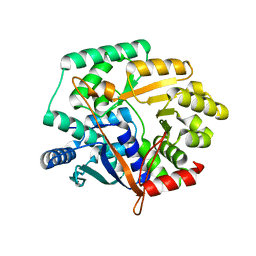 | | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas Gingivalis | | Descriptor: | Nicotinate phosphoribosyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-03 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Nicotinate phosphoribosyltransferase from Porphyromonas gingivalis
To be Published
|
|
2HXU
 
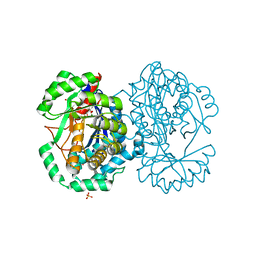 | | Crystal structure of K220A mutant of L-Fuconate Dehydratase from Xanthomonas campestris liganded with Mg++ and L-fuconate | | Descriptor: | 6-deoxy-L-galactonic acid, L-fuconate dehydratase, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: l-Fuconate Dehydratase from Xanthomonas campestris.
Biochemistry, 45, 2006
|
|
7MSG
 
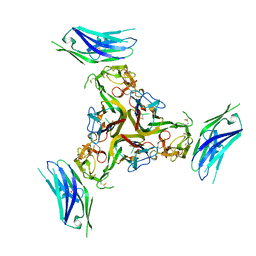 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
1YWK
 
 | |
2HNE
 
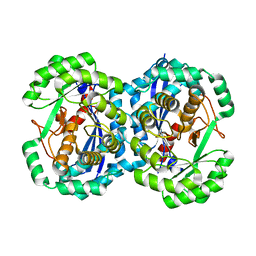 | | Crystal structure of l-fuconate dehydratase from xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-07-12 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of l-fuconate dehydratase from xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
2DW6
 
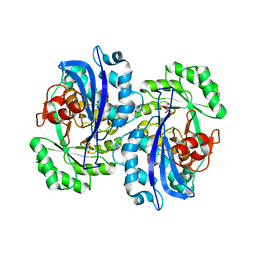 | | Crystal structure of the mutant K184A of D-Tartrate Dehydratase from Bradyrhizobium japonicum complexed with Mg++ and D-tartrate | | Descriptor: | Bll6730 protein, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-07 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: d-Tartrate Dehydratase from Bradyrhizobium japonicum
Biochemistry, 45, 2006
|
|
4I4C
 
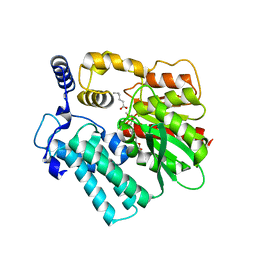 | | Crystal structure of the protein frsA complexed with unknown ligand | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, HEXANOIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Richards, N, Almo, S.C. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational, structural, and kinetic evidence that Vibrio vulnificus FrsA is not a cofactor-independent pyruvate decarboxylase.
Biochemistry, 52, 2013
|
|
3BG2
 
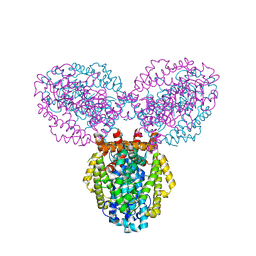 | | Crystal structure of deoxyguanosinetriphosphate triphosphohydrolase from Flavobacterium sp. MED217 | | Descriptor: | DGTP triphosphohydrolase | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the deoxyguanosinetriphosphate triphosphohydrolase from Flavobacterium sp. MED217.
To be Published
|
|
2ZAY
 
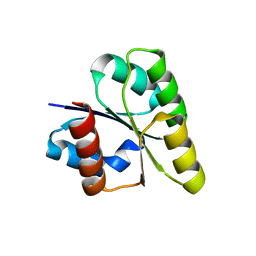 | | Crystal structure of response regulator from Desulfuromonas acetoxidans | | Descriptor: | Response regulator receiver protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of response regulator from Desulfuromonas acetoxidans.
To be Published
|
|
4NG3
 
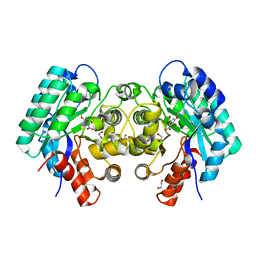 | | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-carboxyvanillate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-01 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with 4-Hydroxy-3-methoxy-5-nitrobenzoic acid
To be Published
|
|
7MSJ
 
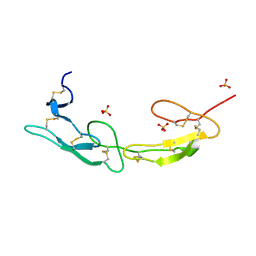 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
1WHO
 
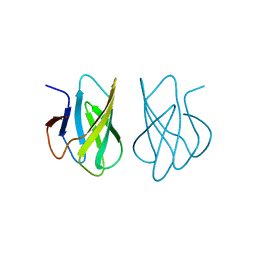 | | ALLERGEN PHL P 2 | | Descriptor: | ALLERGEN PHL P 2 | | Authors: | Fedorov, A.A, Fedorov, E.V, Dolecek, C, Susani, M, Valenta, R, Almo, S.C. | | Deposit date: | 1997-04-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Phl P 2, a Major Timothy Grass (Phleum Pratense) Pollen Allergen
To be Published
|
|
1WVI
 
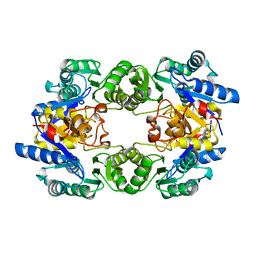 | |
1WV2
 
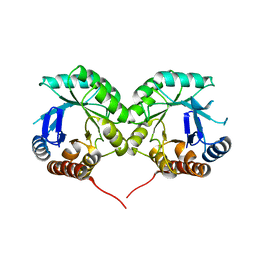 | |
1WHP
 
 | | ALLERGEN PHL P 2 | | Descriptor: | ALLERGEN PHL P 2 | | Authors: | Fedorov, A.A, Fedorov, E.V, Dolecek, C, Susani, M, Valenta, R, Almo, S.C. | | Deposit date: | 1997-04-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Phl P 2, a Major Timothy Grass (Phleum Pratense) Pollen Allergen
To be Published
|
|
1YS9
 
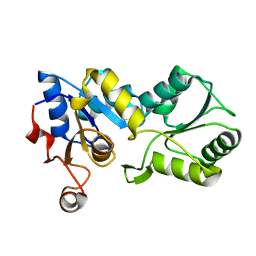 | |
1YRZ
 
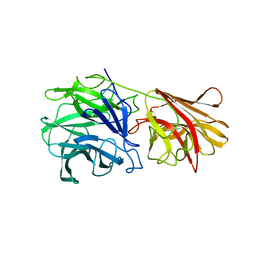 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
1YMY
 
 | | Crystal Structure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the N-acetylglucosamine-6-phosphate deacetylase from Escherichia Coli
To be Published
|
|
1Z4E
 
 | |
4NI8
 
 | | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis complexed with Mn and 5-methoxyisophtalic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-carboxyvanillate decarboxylase, 5-methoxybenzene-1,3-dicarboxylic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase LigW from Sphingomonas paucimobilis complexed with Mn and 5-methoxyisophtalic acid
To be Published
|
|
2HDW
 
 | | Crystal structure of hypothetical protein PA2218 from Pseudomonas Aeruginosa | | Descriptor: | Hypothetical protein PA2218 | | Authors: | Fedorov, A.A, Fedorov, E.V, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein PA2218
from Pseudomonas Aeruginosa
To be Published
|
|
