3DZB
 
 | |
3G1W
 
 | |
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | 分子名称: | ABC transporter, substrate binding protein, GLYCEROL, ... | | 著者 | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-08-25 | | 公開日 | 2008-09-09 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
1T3A
 
 | | Crystal structure of Clostridium botulinum neurotoxin type E catalytic domain | | 分子名称: | CHLORIDE ION, ZINC ION, neurotoxin type E | | 著者 | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | 登録日 | 2004-04-26 | | 公開日 | 2004-06-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
1T3C
 
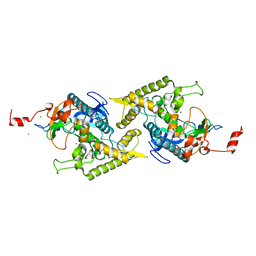 | | Clostridium botulinum type E catalytic domain E212Q mutant | | 分子名称: | CHLORIDE ION, ZINC ION, neurotoxin type E | | 著者 | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | 登録日 | 2004-04-26 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
3GPK
 
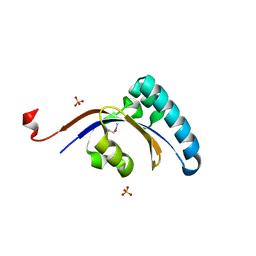 | |
3FFZ
 
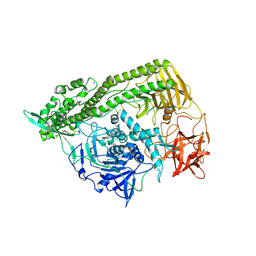 | | Domain organization in Clostridium butulinum neurotoxin type E is unique: Its implication in faster translocation | | 分子名称: | ACETATE ION, Botulinum neurotoxin type E, SODIUM ION, ... | | 著者 | Kumaran, D, Eswaramoorthy, S, Swaminathan, S. | | 登録日 | 2008-12-04 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation.
J.Mol.Biol., 386, 2009
|
|
3H49
 
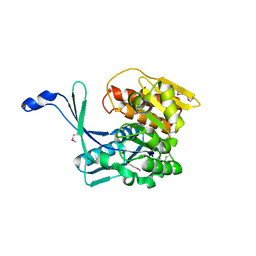 | |
3HDP
 
 | |
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | 分子名称: | Hbl B protein | | 著者 | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-11-02 | | 公開日 | 2006-11-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
2POF
 
 | |
2PLG
 
 | |
2POZ
 
 | |
2Q09
 
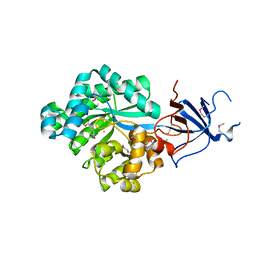 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | 分子名称: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | 著者 | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-05-21 | | 公開日 | 2007-06-05 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
2NXO
 
 | |
2NYG
 
 | |
2PBE
 
 | |
2OOF
 
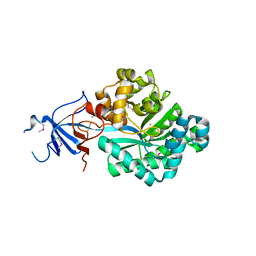 | |
2NN4
 
 | | Crystal structure of Bacillus subtilis yqgQ, Pfam DUF910 | | 分子名称: | Hypothetical protein yqgQ | | 著者 | Damodharan, L, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-10-23 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of YqgQ protein from Bacillus subtilis, a conserved hypothetical protein.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2QVG
 
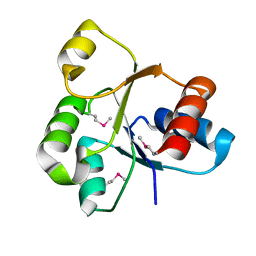 | |
2PUZ
 
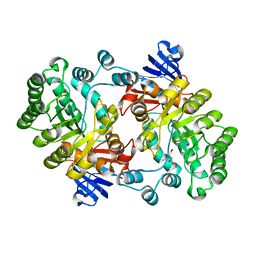 | | Crystal structure of Imidazolonepropionase from Agrobacterium tumefaciens with bound product N-formimino-L-Glutamate | | 分子名称: | CHLORIDE ION, FE (III) ION, Imidazolonepropionase, ... | | 著者 | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-05-09 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 A resolution.
Proteins, 69, 2007
|
|
