3CZT
 
 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 9 | | Descriptor: | CALCIUM ION, Protein S100-B, ZINC ION | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-04-30 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
3D0Y
 
 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 6.5 | | Descriptor: | CALCIUM ION, Protein S100-B, TETRAETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
8AK3
 
 | |
8AK2
 
 | | Drosophila melanogaster UNC89 Protein Kinase Domain 1 (apo) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Obscurin | | Authors: | Dorendorf, T, Zacharchenko, T, Mayans, O. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PK1 from Drosophila obscurin is an inactive pseudokinase with scaffolding properties.
Open Biology, 13, 2023
|
|
8BVM
 
 | |
8BVJ
 
 | | Hfq-Crc-estA translation repression complex | | Descriptor: | Catabolite repression control protein, RNA-binding protein Hfq, estA mRNA | | Authors: | Dendooven, T, Luisi, B.F. | | Deposit date: | 2022-12-04 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Translational regulation by Hfq-Crc assemblies emerges from polymorphic ribonucleoprotein folding.
Embo J., 42, 2023
|
|
8BVH
 
 | |
7OGM
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ)-Hfq | | Descriptor: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase, RNA-binding protein Hfq | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7OGL
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. apo-PNPase | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7OGK
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ) | | Descriptor: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
2H61
 
 | | X-ray structure of human Ca2+-loaded S100B | | Descriptor: | CALCIUM ION, Protein S100-B, TETRAETHYLENE GLYCOL | | Authors: | Ostendorp, T, Heizmann, C.W, Kroneck, P.M.H, Fritz, G. | | Deposit date: | 2006-05-30 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insights into RAGE activation by multimeric S100B.
Embo J., 26, 2007
|
|
8QV0
 
 | | Structure of the native microtubule lattice nucleated from the yeast spindle pole body | | Descriptor: | Tubulin alpha-1 chain, Tubulin beta chain | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 2024
|
|
8QV3
 
 | | Structure of the y-Tubulin Small Complex (yTuSC) as part of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, Spindle pole body component 110, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 2024
|
|
8QV2
 
 | | Structure of the native y-Tubulin Ring Complex (yTuRC) capping microtubule minus ends at the spindle pole body | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component, ... | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 2024
|
|
3D10
 
 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 10.0 | | Descriptor: | CALCIUM ION, Protein S100-B, TRIETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
2RQ8
 
 | | Solution NMR structure of titin I27 domain mutant | | Descriptor: | Titin | | Authors: | Yagawa, K, Oguro, T, Momose, T, Kawano, S, Sato, T, Endo, T. | | Deposit date: | 2009-03-05 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for unfolding pathway-dependent stability of proteins: Vectorial unfolding vs. global unfolding
Protein Sci., 2010
|
|
1EOQ
 
 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: C-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-23 | | Release date: | 2000-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
5TNT
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, N-[(1S,4S,7R)-2-(3-amino-4-chloro[1,2]oxazolo[5,4-c]pyridin-7-yl)-2-azabicyclo[2.2.1]heptan-7-yl]-2-chloro-4-(3-methyl-1H-1,2,4-triazol-1-yl)benzamide, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, A, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TNO
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, SODIUM ION, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, H, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6JNF
 
 | | Cryo-EM structure of the translocator of the outer mitochondrial membrane | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Araiso, Y, Tsutsumi, A, Suzuki, J, Yunoki, K, Kawano, S, Kikkawa, M, Endo, T. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure of the mitochondrial import gate reveals distinct preprotein paths.
Nature, 575, 2019
|
|
7BTW
 
 | | The mitochondrial SAM complex from S.cere | | Descriptor: | Mitochondrial outer membrane beta-barrel protein, SAM37 isoform 1, Sorting assembly machinery 35 kDa subunit | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTX
 
 | | The mitochondrial SAM-Mdm10 supercomplex in GDN micelle from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
7BTY
 
 | | The mitochondrial SAM-Mdm10 supercomplex in Nanodisc from S.cere | | Descriptor: | MDM10 isoform 1, Mitochondrial outer membrane beta-barrel protein, Sorting assembly machinery 35 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Nishizawa, T, Nureki, O, Kikkawa, M, Endo, T. | | Deposit date: | 2020-04-03 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Nature, 590, 2021
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
1OM2
 
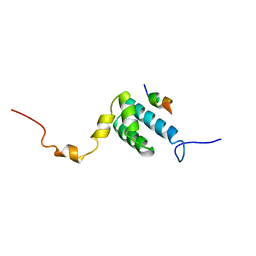 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
