6OA0
 
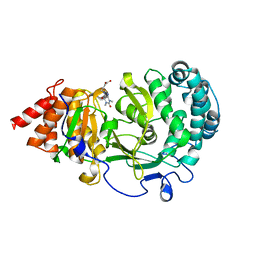 | | Structure of human PARG complexed with JA2-9 | | Descriptor: | 4-(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)butanoic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
1DIZ
 
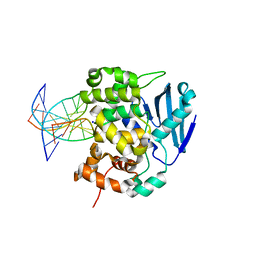 | | CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, DNA (5'-D(*GP*AP*CP*AP*TP*GP*AP*(NRI)P*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Hollis, T, Ichikawa, Y, Ellenberger, T.E. | | Deposit date: | 1999-11-30 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA.
EMBO J., 19, 2000
|
|
1BNK
 
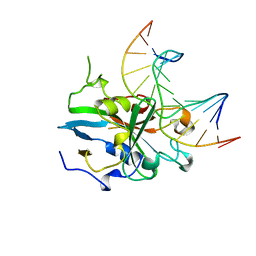 | | HUMAN 3-METHYLADENINE DNA GLYCOSYLASE COMPLEXED TO DNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*YRRP*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), PROTEIN (3-METHYLADENINE DNA GLYCOSYLASE) | | Authors: | Lau, A.Y, Schaerer, O.D, Samson, L, Verdine, G.L, Ellenberger, T. | | Deposit date: | 1998-07-29 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision.
Cell(Cambridge,Mass.), 95, 1998
|
|
2HJH
 
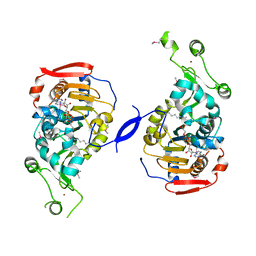 | | Crystal Structure of the Sir2 deacetylase | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, NAD-dependent histone deacetylase SIR2, NICOTINAMIDE, ... | | Authors: | Hall, B.E, Ellenberger, T.E. | | Deposit date: | 2006-06-30 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Autoregulation of the yeast Sir2 deacetylase by reaction and trapping of a pseudosubstrate motif in the active site
To be Published
|
|
2IQC
 
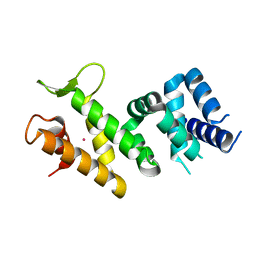 | | Crystal structure of Human FancF Protein that Functions in the Assembly of a DNA Damage Signaling Complex | | Descriptor: | Fanconi anemia group F protein, MERCURY (II) ION | | Authors: | Kowal, P, Gurtan, A.M, Stuckert, P, Lehmann, C, D'Andrea, A, Ellenberger, T.E. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex.
J.Biol.Chem., 282, 2007
|
|
1CR1
 
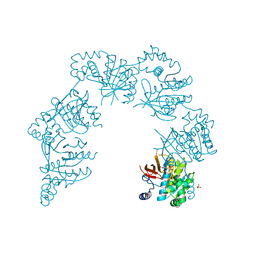 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR4
 
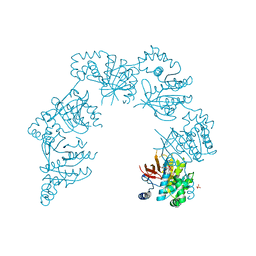 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR2
 
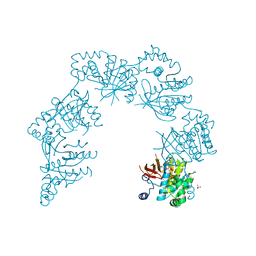 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1AE9
 
 | |
1NUI
 
 | | Crystal Structure of the primase fragment of Bacteriophage T7 primase-helicase protein | | Descriptor: | DNA primase/helicase, MAGNESIUM ION, ZINC ION | | Authors: | Kato, M, Ito, T, Wagner, G, Richardson, C.C, Ellenberger, T. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modular Architecture of the Bacteriophage T7 Primase Couples RNA primer Synthesis to DNA Synthesis
Mol.Cell, 11, 2003
|
|
1P7D
 
 | | Crystal structure of the Lambda Integrase (residues 75-356) bound to DNA | | Descriptor: | 26-MER, 5'-D(*CP*AP*AP*TP*GP*CP*CP*AP*AP*CP*TP*TP*T)-3', Integrase | | Authors: | Aihara, H, Kwon, H.J, Nunes-Duby, S.E, Landy, A, Ellenberger, T. | | Deposit date: | 2003-05-01 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Conformational Switch Controls the DNA Cleavage Activity of Lambda Integrase
Mol.Cell, 12, 2003
|
|
1X9S
 
 | | T7 DNA polymerase in complex with a primer/template DNA containing a disordered N-2 aminofluorene on the template, crystallized with dideoxy-CTP as the incoming nucleotide. | | Descriptor: | 5'-D(*CP*CP*CP*(AFG)P*AP*TP*CP*AP*CP*AP*CP*TP*AP*CP*CP*AP*AP*TP*CP*AP*CP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*GP*TP*GP*AP*TP*T*GP*GP*TP*AP*GP*TP*GP*TP*GP*AP*(2DT))-3', DNA polymerase, ... | | Authors: | Dutta, S, Li, Y, Johnson, D, Dzantiev, L, Richardson, C.C, Romano, L.J, Ellenberger, T. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1F6O
 
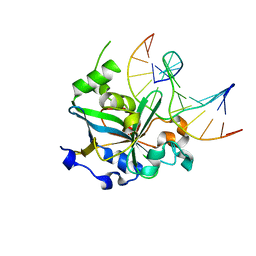 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH DNA | | Descriptor: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(YRR)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | Deposit date: | 2000-06-22 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F4R
 
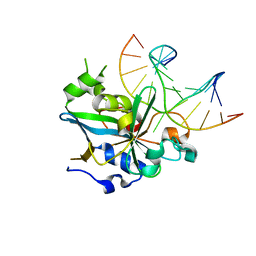 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH 1,N6-ETHENOADENINE-DNA | | Descriptor: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDA)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | Authors: | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | Deposit date: | 2000-06-08 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1TK8
 
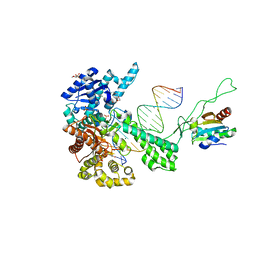 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and dAMP at the elongation site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*CP*CP*CP*AP*(8OG)P*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3', ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
1NYH
 
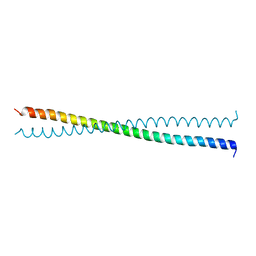 | | Crystal Structure of the Coiled-coil Dimerization Motif of Sir4 | | Descriptor: | Regulatory protein SIR4 | | Authors: | Chang, J.F, Hall, B.E, Tanny, J.C, Moazed, D, Filman, D, Ellenberger, T. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Coiled-coil Dimerization Motif of Sir4 and Its Interaction With Sir3
Structure, 11, 2003
|
|
1CR0
 
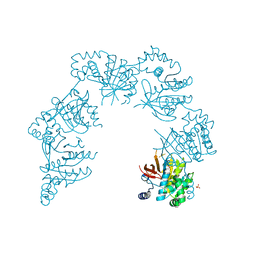 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE4 PROTEIN OF BACTERIOPHAGE T7 | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1MPG
 
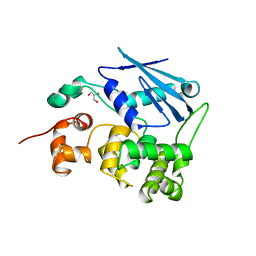 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
1PU6
 
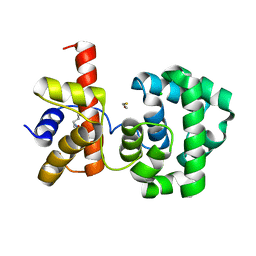 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-METHYLADENINE DNA GLYCOSYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1TKD
 
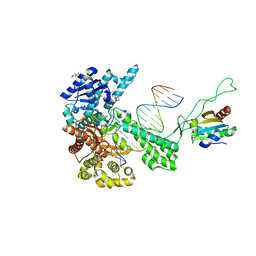 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and dCMP at the elongation site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*CP*CP*CP*AP*(8OG)P*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
1PU7
 
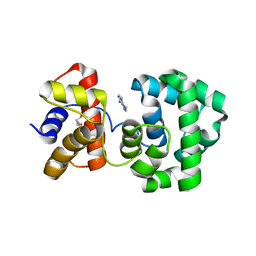 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1D5Y
 
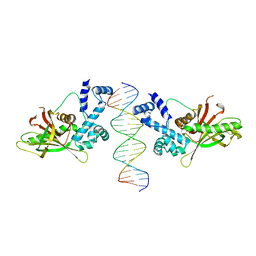 | | CRYSTAL STRUCTURE OF THE E. COLI ROB TRANSCRIPTION FACTOR IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*CP*AP*TP*TP*CP*AP*GP*TP*GP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*GP*AP*AP*TP*GP*TP*CP*AP*AP*AP*G)-3'), ROB TRANSCRIPTION FACTOR | | Authors: | Kwon, H.J, Bennik, M.H.J, Demple, B, Ellenberger, T. | | Deposit date: | 1999-10-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA.
Nat.Struct.Biol., 7, 2000
|
|
1Z1B
 
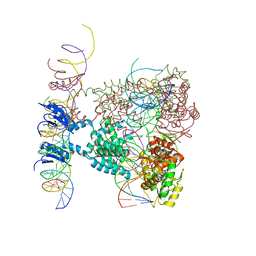 | | Crystal structure of a lambda integrase dimer bound to a COC' core site | | Descriptor: | 26-MER DNA, 29-MER DNA, 5'-D(*CP*T*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
1PU8
 
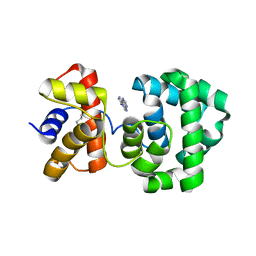 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1Q57
 
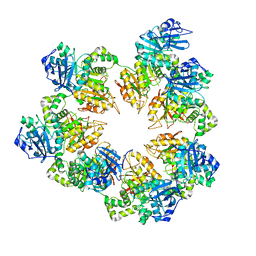 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
