2KQX
 
 | |
1D5G
 
 | |
4LRY
 
 | | Crystal Structure of the E.coli DhaR(N)-DhaK(T79L) complex | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase operon regulatory protein, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
3UCS
 
 | |
1Y7X
 
 | | Solution structure of a two-repeat fragment of major vault protein | | Descriptor: | Major vault protein | | Authors: | Kozlov, G, Vavelyuk, O, Minailiuc, O, Banville, D, Gehring, K, Ekiel, I. | | Deposit date: | 2004-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-repeat fragment of major vault protein.
J.Mol.Biol., 356, 2006
|
|
2H7A
 
 | | NMR Structure of the Conserved Protein YcgL from Escherichia coli representing the DUF709 Family Reveals a Novel a/b/a Sandwich Fold | | Descriptor: | Hypothetical protein ycgL | | Authors: | Minailiuc, O.M, Vavelyuk, O, Ekiel, I, Hung, M.-Ni, Cygler, M, Gandhi, S, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-06-01 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of YcgL, a conserved protein from Escherichia coli representing the DUF709 family, with a novel alpha/beta/alpha sandwich fold.
Proteins, 66, 2007
|
|
4LRZ
 
 | | Crystal Structure of the E.coli DhaR(N)-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-dependent dihydroxyacetone kinase operon regulatory protein, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
4LRX
 
 | | Crystal Structure of the E.coli DhaR(N)-DhaK complex | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase operon regulatory protein, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Shi, R, McDonald, L, Cygler, M, Ekiel, I. | | Deposit date: | 2013-07-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Coiled-Coil Helix Rotation Selects Repressing or Activating State of Transcriptional Regulator DhaR.
Structure, 22, 2014
|
|
2PQ4
 
 | |
1L1P
 
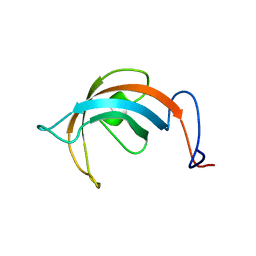 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
1R6H
 
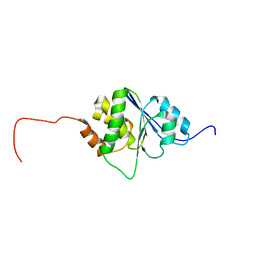 | | Solution Structure of human PRL-3 | | Descriptor: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | Authors: | Kozlov, G, Gehring, K, Ekiel, I. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
1JGN
 
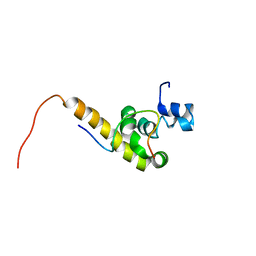 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1SSL
 
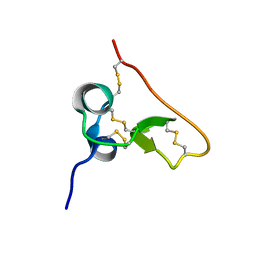 | | Solution structure of the PSI domain from the Met receptor | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Kozlov, G, Perreault, A, Schrag, J.D, Cygler, M, Gehring, K, Ekiel, I. | | Deposit date: | 2004-03-24 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Insights into function of PSI domains from structure of the Met receptor PSI domain.
Biochem.Biophys.Res.Commun., 321, 2004
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
3PDZ
 
 | |
3PNL
 
 | | Crystal Structure of E.coli Dha kinase DhaK-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNO
 
 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) | | Descriptor: | PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNK
 
 | | Crystal Structure of E.coli Dha kinase DhaK | | Descriptor: | GLYCEROL, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNQ
 
 | | Crystal Structure of E.coli Dha kinase DhaK (H56N) complex with Dha | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PNM
 
 | | Crystal Structure of E.coli Dha kinase DhaK (H56A) | | Descriptor: | PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1G9L
 
 | | SOLUTION STRUCTURE OF THE PABC DOMAIN OF HUMAN POLY(A) BINDING PROTEIN | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1 | | Authors: | Kozlov, G, Trempe, J.-F, Khaleghpour, K, Kahvejian, A, Ekiel, I, Gehring, K. | | Deposit date: | 2000-11-24 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the C-terminal PABC domain of human poly(A)-binding protein.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IFW
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN OF POLY(A) BINDING PROTEIN FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | POLYADENYLATE-BINDING PROTEIN, CYTOPLASMIC AND NUCLEAR | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Sprules, T, Ekiel, I, Gehring, K. | | Deposit date: | 2001-04-13 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein.
J.Biol.Chem., 277, 2002
|
|
1GH9
 
 | |
1GH8
 
 | |
