1Z6K
 
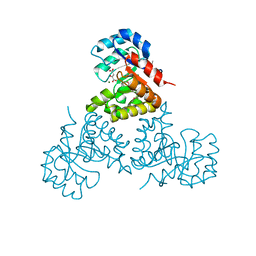 | | Citrate lyase beta subunit complexed with oxaloacetate and magnesium from M. tuberculosis | | Descriptor: | Citrate Lyase beta subunit, MAGNESIUM ION, OXALOACETATE ION | | Authors: | Goulding, C.W, Bowers, P.M, Segelke, B, Lekin, T, Kim, C.Y, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure and computational analysis of Mycobacterium tuberculosis protein CitE suggest a novel enzymatic function.
J.Mol.Biol., 365, 2007
|
|
6OIZ
 
 | | Amyloid-Beta (20-34) wild type | | Descriptor: | Amyloid beta A4 protein | | Authors: | Sawaya, M.R, Warmack, R.A, Zee, C.T, Gonen, T, Clarke, S.G, Eisenberg, D.S. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Structure of amyloid-beta (20-34) with Alzheimer's-associated isomerization at Asp23 reveals a distinct protofilament interface.
Nat Commun, 10, 2019
|
|
3DG1
 
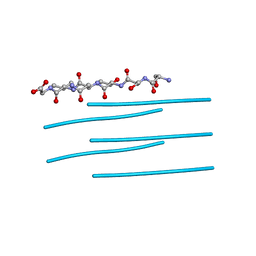 | | Segment SSTNVG derived from IAPP | | Descriptor: | SSTNVG from Islet Amyloid Polypeptide | | Authors: | Wiltzius, J.J, Sievers, S.A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin).
Protein Sci., 17, 2008
|
|
3DGJ
 
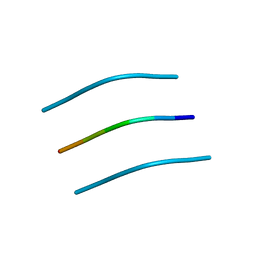 | | NNFGAIL segment from Islet Amyloid Polypeptide (IAPP or amylin) | | Descriptor: | NNFGAIL peptide | | Authors: | Wiltzius, J.J, Sievers, S.A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2008-06-13 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin).
Protein Sci., 17, 2008
|
|
1LGR
 
 | |
1F1G
 
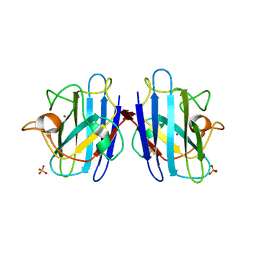 | | Crystal structure of yeast cuznsod exposed to nitric oxide | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, PHOSPHATE ION, ... | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-12 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F18
 
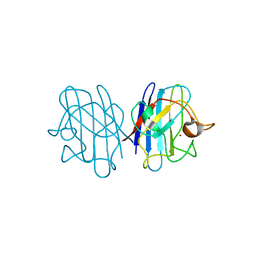 | | Crystal structure of yeast copper-zinc superoxide dismutase mutant GLY85ARG | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F0P
 
 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B WITH TREHALOSE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85-B, ... | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
1F1A
 
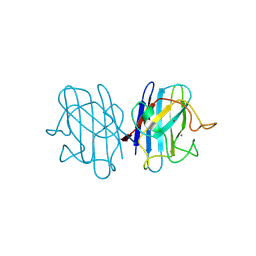 | | Crystal structure of yeast H48Q cuznsod fals mutant analog | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F0N
 
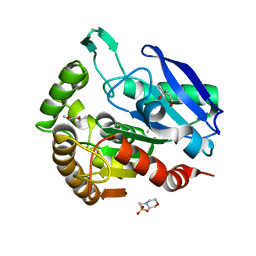 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85B | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
1F1D
 
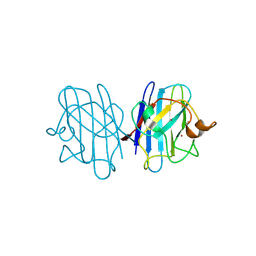 | | Crystal structure of yeast H46C cuznsod mutant | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F0X
 
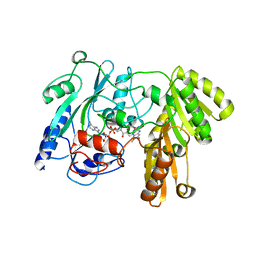 | | CRYSTAL STRUCTURE OF D-LACTATE DEHYDROGENASE, A PERIPHERAL MEMBRANE RESPIRATORY ENZYME. | | Descriptor: | D-LACTATE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Dym, O, Pratt, E.A, Ho, C, Eisenberg, D. | | Deposit date: | 2000-05-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1F1H
 
 | |
1F52
 
 | | CRYSTAL STRUCTURE OF GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM CO-CRYSTALLIZED WITH ADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE SYNTHETASE, ... | | Authors: | Gill, H.S, Pfluegl, G.M.U, Eisenberg, D. | | Deposit date: | 2000-06-12 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition.
Biochemistry, 40, 2001
|
|
3FPO
 
 | |
4QX0
 
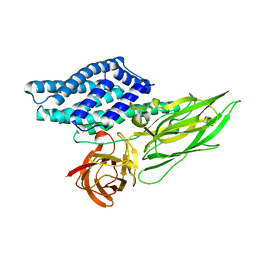 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1FPY
 
 | |
4QX3
 
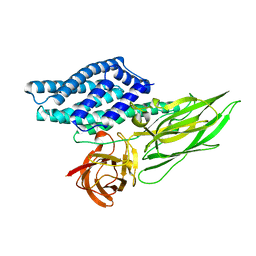 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3FTH
 
 | |
1YJO
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
1YJP
 
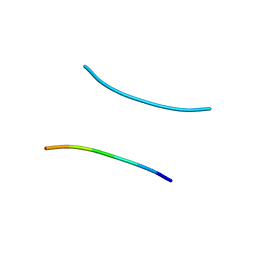 | | Structure of GNNQQNY from yeast prion Sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
1JS0
 
 | | Crystal Structure of 3D Domain-swapped RNase A Minor Trimer | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Gotte, G, Libonati, M, Eisenberg, D. | | Deposit date: | 2001-08-15 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the two 3D domain-swapped RNase A trimers.
Protein Sci., 11, 2002
|
|
1YKW
 
 | |
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3DMK
 
 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
