2LHS
 
 | |
7Z65
 
 | | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, GH18 family | | Authors: | Madhuprakash, J, Dalhus, B, Rohr, A.K, Bissaro, B, Vaaje-Kolstad, G, Sorlie, M, Eijsink, V.G. | | Deposit date: | 2022-03-11 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.108 Å) | | Cite: | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases
To Be Published
|
|
7Z64
 
 | | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Madhuprakash, J, Dalhus, B, Rohr, A.K, Bissaro, B, Vaaje-Kolstad, G, Sorlie, M, Eijsink, V.G. | | Deposit date: | 2022-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases
To Be Published
|
|
7ZJB
 
 | | Structural and functional characterization of the bacterial lytic polysaccharide Monooxygenase ScLPMO10D | | Descriptor: | COPPER (II) ION, Putative secreted cellulose-binding protein, SODIUM ION, ... | | Authors: | Votvik, A.K, Rohr, A.K, Stepnov, A.A, Bissaro, B, Sorlie, M, Eijsink, V.G.H, Forsberg, Z. | | Deposit date: | 2022-04-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and functional characterization of the catalytic domain of a cell-wall anchored bacterial lytic polysaccharide monooxygenase from Streptomyces coelicolor.
Sci Rep, 13, 2023
|
|
1W1Y
 
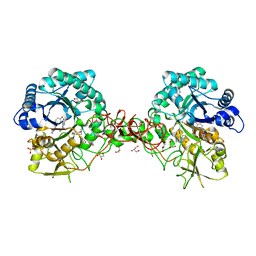 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Tyr-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-TYROSINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
7P3U
 
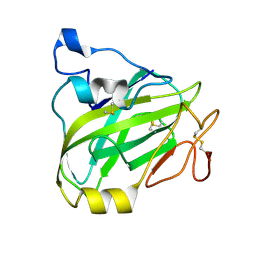 | | Chitin-active fungal AA11 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, putative | | Authors: | Rohr, A.K, Stoepamo, F.G, Eijsink, V.G.H. | | Deposit date: | 2021-07-08 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of a lytic polysaccharide monooxygenase from Aspergillus fumigatus shows functional variation among family AA11 fungal LPMOs.
J.Biol.Chem., 297, 2021
|
|
5OPF
 
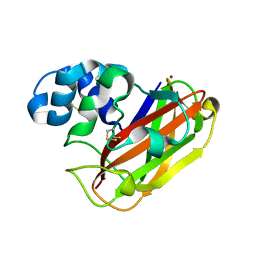 | | Structure of LPMO10B from from Micromonospora aurantiaca | | Descriptor: | COPPER (II) ION, Chitin-binding domain 3 protein | | Authors: | Forsberg, Z, Bissaro, B, Gullesen, J, Dalhus, B, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.081 Å) | | Cite: | Structural determinants of bacterial lytic polysaccharide monooxygenase functionality.
J. Biol. Chem., 293, 2018
|
|
2XWX
 
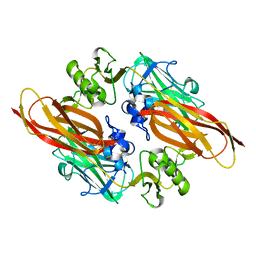 | | Vibrio cholerae colonization factor GbpA crystal structure | | Descriptor: | GLCNAC-BINDING PROTEIN A | | Authors: | Wong, E, Vaaje-Kolstad, G, Ghosh, A, Guerrero, R.H, Konarev, P.V, Ibrahim, A.F.M, Svergun, D.I, Eijsink, V.G.H, Chatterjee, N.S, van Aalten, D.M.F. | | Deposit date: | 2010-11-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio Cholerae Colonization Factor Gbpa Possesses a Modular Structure that Governs Binding to Different Host Surfaces.
Plos Pathog., 8, 2012
|
|
1OGB
 
 | | Chitinase b from Serratia marcescens mutant D142N | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-29 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
4YHG
 
 | | NATIVE BACTEROIDETES-AFFILIATED GH5 CELLULASE LINKED WITH A POLYSACCHARIDE UTILIZATION LOCUS | | Descriptor: | GH5, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Naas, A.E, MacKenzie, A.K, Dalhus, B, Eijsink, V.G.H, Pope, P.B. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Features of a Bacteroidetes-Affiliated Cellulase Linked with a Polysaccharide Utilization Locus.
Sci Rep, 5, 2015
|
|
4YHE
 
 | | NATIVE BACTEROIDETES-AFFILIATED GH5 CELLULASE LINKED WITH A POLYSACCHARIDE UTILIZATION LOCUS | | Descriptor: | GH5 | | Authors: | Naas, A.E, MacKenzie, A.K, Dalhus, B, Eijsink, V.G.H, Pope, P.B. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Features of a Bacteroidetes-Affiliated Cellulase Linked with a Polysaccharide Utilization Locus.
Sci Rep, 5, 2015
|
|
1OGG
 
 | | chitinase b from serratia marcescens mutant d142n in complex with inhibitor allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE B, ... | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Rao, F.V, Peter, M.G, Synstad, B, van Aalten, D.M.F, Eijsink, V.G.H. | | Deposit date: | 2003-04-30 | | Release date: | 2004-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the D142N Mutant of the Family 18 Chitinase Chib from Serratia Marcescens and its Complex with Allosamidin
Biochim.Biophys.Acta, 1696, 2004
|
|
6YHH
 
 | | X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylglucosaminidase-like protein Glycoside hydrolase family 20, GLYCEROL | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V.G.H, Pope, P.B, Branden, G, Larsbrink, J. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
1E6Z
 
 | | CHITINASE B FROM SERRATIA MARCESCENS WILDTYPE IN COMPLEX WITH CATALYTIC INTERMEDIATE | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-23 | | Release date: | 2001-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1E6P
 
 | | Chitinase B from Serratia marcescens inactive mutant E144Q | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-22 | | Release date: | 2001-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1O6I
 
 | | Chitinase B from Serratia marcescens complexed with the catalytic intermediate mimic cyclic dipeptide CI4. | | Descriptor: | Chitinase, GLYCEROL, SULFATE ION, ... | | Authors: | Houston, D.R, Eggleston, I, Synstad, B, Eijsink, V.G.H, van Aalten, D.M.F. | | Deposit date: | 2002-10-03 | | Release date: | 2003-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The cyclic dipeptide CI-4 [cyclo-(l-Arg-d-Pro)] inhibits family 18 chitinases by structural mimicry of a reaction intermediate.
Biochem. J., 368, 2002
|
|
6FAO
 
 | | Discovery and characterization of a thermostable GH6 endoglucanase from a compost metagenome | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 6, SULFATE ION | | Authors: | Jensen, M.S, Fredriksen, L, MacKenzie, A.K, Pope, P.B, Chylenski, P, Leiros, I, Williamson, A.K, Christopeit, T, Ostby, H, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome.
PLoS ONE, 13, 2018
|
|
6F8N
 
 | | Key residues affecting transglycosylation activity in family 18 chitinases - Insights into donor and acceptor subsites | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Madhuprakash, J, Dalhus, B, Swaroopa Rani, T, Podile, A.R, Eijsink, V.G.H, Sorlie, M. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Key Residues Affecting Transglycosylation Activity in Family 18 Chitinases: Insights into Donor and Acceptor Subsites.
Biochemistry, 57, 2018
|
|
6HM1
 
 | | Structural and thermodynamic signatures of ligand binding to an enigmatic chitinase-D from Serratia proteamaculans | | Descriptor: | 1,2-ETHANEDIOL, ALLOSAMIDIN, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Dalhus, B, Vaaje-Kolstad, G, Eijsink, V.G.H, Sorlie, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Thermodynamic Signatures of Ligand Binding to the Enigmatic Chitinase D of Serratia proteamaculans.
J.Phys.Chem.B, 123, 2019
|
|
4OY6
 
 | | Structure of ScLPMO10B in complex with copper. | | Descriptor: | ACETATE ION, COPPER (II) ION, Putative secreted cellulose-binding protein, ... | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OY7
 
 | | Structure of cellulose active LPMO CelS2 (ScLPMO10C) in complex with Copper. | | Descriptor: | CALCIUM ION, COPPER (II) ION, Putative secreted cellulose binding protein | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OY8
 
 | | Structure of ScLPMO10B in complex with zinc. | | Descriptor: | ACETATE ION, Putative secreted cellulose-binding protein, ZINC ION | | Authors: | Forsberg, Z, Mackenzie, A.K, Sorlie, M, Rohr, A.K, Helland, R, Arvai, A.S, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2014-02-11 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1GOI
 
 | | Crystal structure of the D140N mutant of chitinase B from Serratia marcescens at 1.45 A resolution | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Kolstad, G, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2001-10-21 | | Release date: | 2001-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the D140N Mutant of Chitinase B from Serratia Marcescens at 1.45 A Resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4A02
 
 | | X-ray crystallographic structure of EfCBM33A | | Descriptor: | CHITIN BINDING PROTEIN | | Authors: | Vaaje-Kolstad, G, Bohle, L.A, Gaseidnes, S, Dalhus, B, Bjoras, M, Mathiesen, G, Eijsink, V.G.H. | | Deposit date: | 2011-09-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Characterization of the Chitinolytic Machinery of Enterococcus Faecalis V583 and High Resolution Structure of its Oxidative Cbm33 Enzyme
J.Mol.Biol., 416, 2012
|
|
5FJQ
 
 | | Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus | | Descriptor: | CARBOHYDRATE BINDING PROTEIN, PUTATIVE, CPB33A, ... | | Authors: | Forsberg, Z, Nelson, C.E, Dalhus, B, Mekasha, S, Loose, J.S.M, Rohr, A.K, Eijsink, V.G.H, Gardner, J.G, Vaaje-Kolstad, G. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio Japonicus
J.Biol.Chem., 291, 2016
|
|
