5VKZ
 
 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
4O32
 
 | | Structure of a malarial protein | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Egea, P.F, Koehl, A, Peng, M, Cascio, D. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystal structure and solution characterization of the thioredoxin-2 from Plasmodium falciparum, a constituent of an essential parasitic protein export complex.
Biochem.Biophys.Res.Commun., 456, 2015
|
|
3MP7
 
 | |
3P5S
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Stauffler, H, Kohn, I, Cakou-Kefir, C, Stroud, R.M, Kellenberburger, E, Schuber, F. | | Deposit date: | 2010-10-10 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
4XBI
 
 | | Structure Of A Malarial Protein Involved in Proteostasis | | Descriptor: | ClpB protein, putative,Green fluorescent protein, SULFATE ION | | Authors: | Egea, P.F, Ah Young, A.P, Cascio, D. | | Deposit date: | 2014-12-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural mapping of the ClpB ATPases of Plasmodium falciparum: Targeting protein folding and secretion for antimalarial drug design.
Protein Sci., 24, 2015
|
|
3KOU
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-11-13 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
4IRF
 
 | |
4IOD
 
 | |
1FBY
 
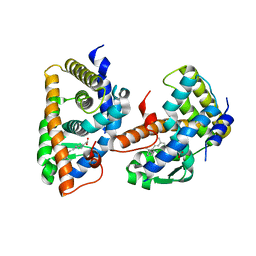 | | CRYSTAL STRUCTURE OF THE HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Egea, P.F, Mitschler, A, Rochel, N, Ruff, M, Chambon, P, Moras, D. | | Deposit date: | 2000-07-17 | | Release date: | 2000-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid.
EMBO J., 19, 2000
|
|
1MZN
 
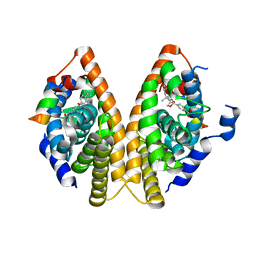 | | CRYSTAL STRUCTURE at 1.9 ANGSTROEMS RESOLUTION OF THE HOMODIMER OF HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO THE SYNTHETIC AGONIST COMPOUND BMS 649 AND A COACTIVATOR PEPTIDE | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-10-09 | | Release date: | 2002-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
1RJ9
 
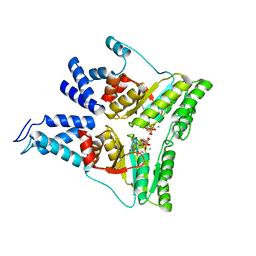 | | Structure of the heterodimer of the conserved GTPase domains of the Signal Recognition Particle (Ffh) and Its Receptor (FtsY) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Signal Recognition Protein, ... | | Authors: | Egea, P.F, Shan, S.O, Napetschnig, J, Savage, D.F, Walter, P, Stroud, R.M. | | Deposit date: | 2003-11-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate twinning activates the signal recognition particle and its receptor
Nature, 427, 2004
|
|
1MVC
 
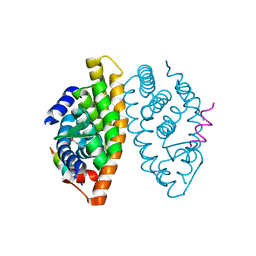 | | Crystal structure of the human RXR alpha ligand binding domain bound to the synthetic agonist compound BMS 649 and a coactivator peptide | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-24 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
1MV9
 
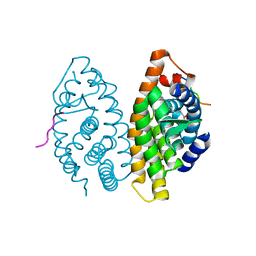 | | Crystal Structure of the human RXR alpha ligand binding domain bound to the eicosanoid DHA (Docosa Hexaenoic Acid) and a coactivator peptide | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-24 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
3E70
 
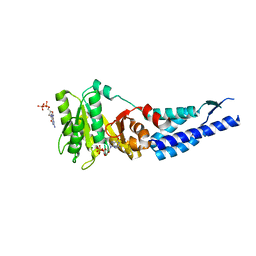 | | Structures and conformations in solution of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus Furiosus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane.
Plos One, 3, 2008
|
|
3DM5
 
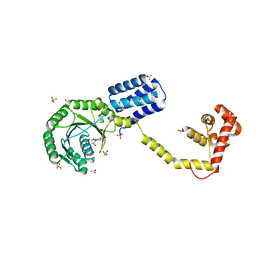 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3DMD
 
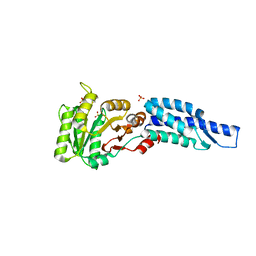 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3DLU
 
 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | BROMIDE ION, MALONATE ION, Signal recognition particle 19 kDa protein | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-29 | | Release date: | 2008-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3GHH
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N.J, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3GC6
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ecto-NAD+ glycohydrolase (CD38 molecule), SULFATE ION | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Oppenheimer, N, Kellenberger, E, Schuber, F. | | Deposit date: | 2009-02-21 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3DM9
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3GH3
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, Ecto-NAD+ glycohydrolase (CD38 molecule), ... | | Authors: | Egea, P.F, Muller-Steffner, H, Stroud, R.M, Kellenberger, E, Oppenheimer, N, Schuber, F. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3DLV
 
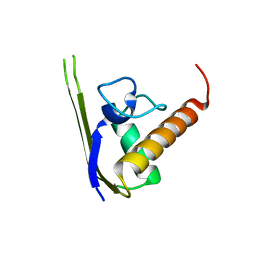 | | Structures of SRP54 and SRP19, the two proteins assembling the ribonucleic core of the Signal Recognition Particle from the archaeon Pyrococcus furiosus. | | Descriptor: | Signal recognition particle 19 kDa protein | | Authors: | Egea, P.F, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-29 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus.
Plos One, 3, 2008
|
|
3BN9
 
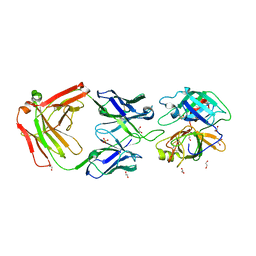 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
4O2X
 
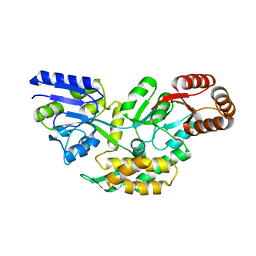 | | Structure of a malarial protein | | Descriptor: | Maltose-binding periplasmic protein, ATP-dependent Clp protease adaptor protein ClpS containing protein chimeric construct | | Authors: | AhYoung, A.P, Koehl, A, Cascio, D, Egea, P.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a putative ClpS N-end rule adaptor protein from the malaria pathogen Plasmodium falciparum.
Protein Sci., 25, 2016
|
|
1RC2
 
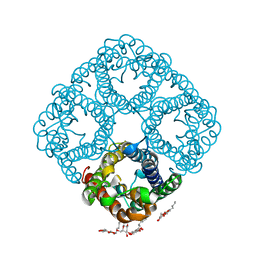 | | 2.5 Angstrom Resolution X-ray Structure of Aquaporin Z | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, Aquaporin Z | | Authors: | Savage, D.F, Egea, P.F, Robles, Y.C, O'Connell III, J.D, Stroud, R.M. | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z
Plos Biol., 1, 2003
|
|
