8B14
 
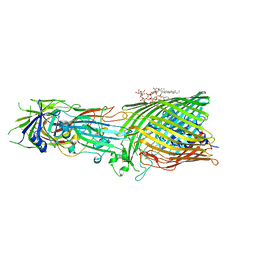 | | T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA | | 分子名称: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, FhuA iron-ferrichrome transporter, [(2R,3S,4R,5R,6R)-2-[[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-4-[(2R,4R,5R,6R)-6-[(1R)-1,2-bis(oxidanyl)ethyl]-2-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-carboxy-5-oxidanyl-oxan-2-yl]oxymethyl]-5-[[(3R)-3-dodecanoyloxytetradecanoyl]amino]-4-(3-nonanoyloxypropanoyloxy)-6-[[(2R,3S,4R,5R,6R)-3-oxidanyl-4-[(3S)-3-oxidanyltetradecanoyl]oxy-5-[[(3R)-3-oxidanyltridecanoyl]amino]-6-phosphonatooxy-oxan-2-yl]methoxy]oxan-3-yl] phosphate, ... | | 著者 | Degroux, S, Effantin, G, Linares, R, Schoehn, G, Breyton, C. | | 登録日 | 2022-09-09 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger.
J.Virol., 97, 2023
|
|
4UPB
 
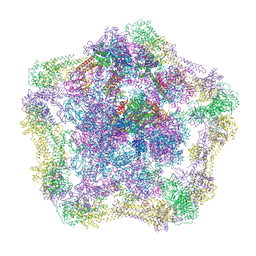 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | 分子名称: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | 著者 | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | 登録日 | 2014-06-15 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
4UPF
 
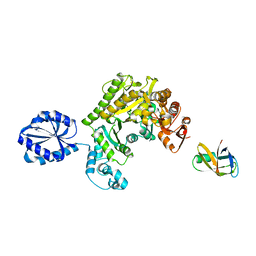 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | 分子名称: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | 著者 | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | 登録日 | 2014-06-16 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.5 Å) | | 主引用文献 | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | 分子名称: | Tetrahedral aminopeptidase, ZINC ION | | 著者 | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | 登録日 | 2019-04-02 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | 主引用文献 | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | 分子名称: | Inducible lysine decarboxylase | | 著者 | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | 登録日 | 2020-04-10 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YN6
 
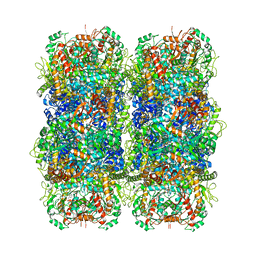 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | 分子名称: | Inducible lysine decarboxylase | | 著者 | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | 登録日 | 2020-04-10 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8A1R
 
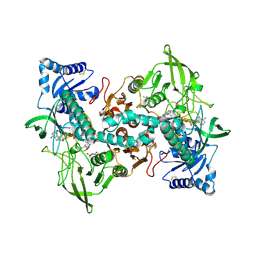 | | cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor | | 分子名称: | (2~{R},3~{R},4~{S},5~{R})-2-[3-[[[(1~{R},2~{R},3~{R},5~{S})-2,6,6-trimethyl-3-bicyclo[3.1.1]heptanyl]amino]methyl]indol-1-yl]oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | 著者 | Ardini, M, Angelucci, F, Fata, F, Gabriele, F, Effantin, G, Ling, W, Williams, D.L, Petukhova, V.Z, Petukhov, P.A. | | 登録日 | 2022-06-01 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Non-covalent inhibitors of thioredoxin glutathione reductase with schistosomicidal activity in vivo.
Nat Commun, 14, 2023
|
|
6Z6G
 
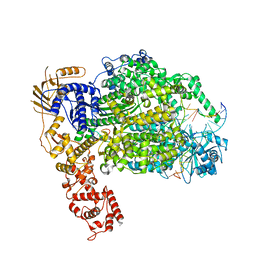 | | Cryo-EM structure of La Crosse virus polymerase at pre-initiation stage | | 分子名称: | 3'vRNA 1-16, 5'vRNA 1-10, 5'vRNA 9-16, ... | | 著者 | Arragain, B, Effantin, G, Gerlach, P, Reguera, J, Schoehn, G, Cusack, S, Malet, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
6Z8K
 
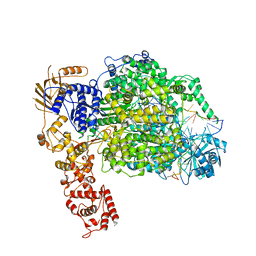 | | La Crosse virus polymerase at elongation mimicking stage | | 分子名称: | La Crosse virus 3' vRNA (1-16), La Crosse virus 5' vRNA (9-16), La Crosse virus 5' vRNA 1-10, ... | | 著者 | Arragain, B, Effantin, G, Schoehn, G, Cusack, S, Malet, H. | | 登録日 | 2020-06-02 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Pre-initiation and elongation structures of full-length La Crosse virus polymerase reveal functionally important conformational changes.
Nat Commun, 11, 2020
|
|
8BCU
 
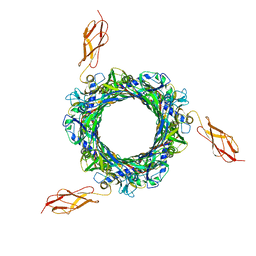 | |
8BCP
 
 | |
6RLP
 
 | |
4D80
 
 | | Metallosphera sedula Vps4 crystal structure | | 分子名称: | AAA ATPASE, CENTRAL DOMAIN PROTEIN | | 著者 | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | 登録日 | 2014-12-02 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
4D82
 
 | | Metallosphera sedula Vps4 crystal structure | | 分子名称: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | 著者 | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | 登録日 | 2014-12-02 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
4D81
 
 | | Metallosphera sedula Vps4 crystal structure | | 分子名称: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | 著者 | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | 登録日 | 2014-12-02 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
